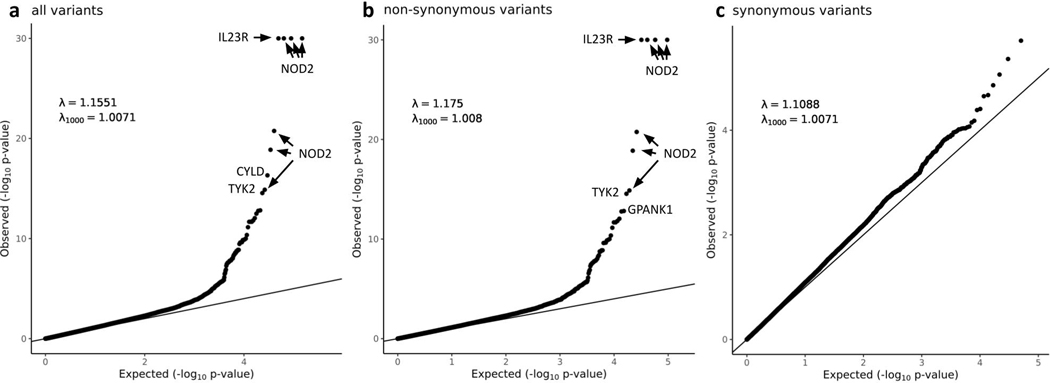
Extended Data Fig. 3. QQ plots of the heterogeneity-of-effect test between the Nextera and Twist discovery cohorts.
Only QC passed variants with minor allele frequency in NFE between 0.0001 and 0.10 were included. a, all variants. b, non-synonymous variants. c, synonymous variants. In a and b, the y axis is capped at -log10 p = 30 while the top four variants (three in NOD2 and one in IL23R) have -log10 p > 100. In c, to remove the synonymous variants that tag causal non-synonymous variants and artifacts through LD, we removed loci hosting large-effect coding variants (IL23R, NOD2, LRRK2, TYK2, ATG16L1, SLC39A8, PTGER4, IRGM, CARD9), implicated by variants removed in the heterogeneous test (AHNAK2, LILRA), and with long range LD (MHC).