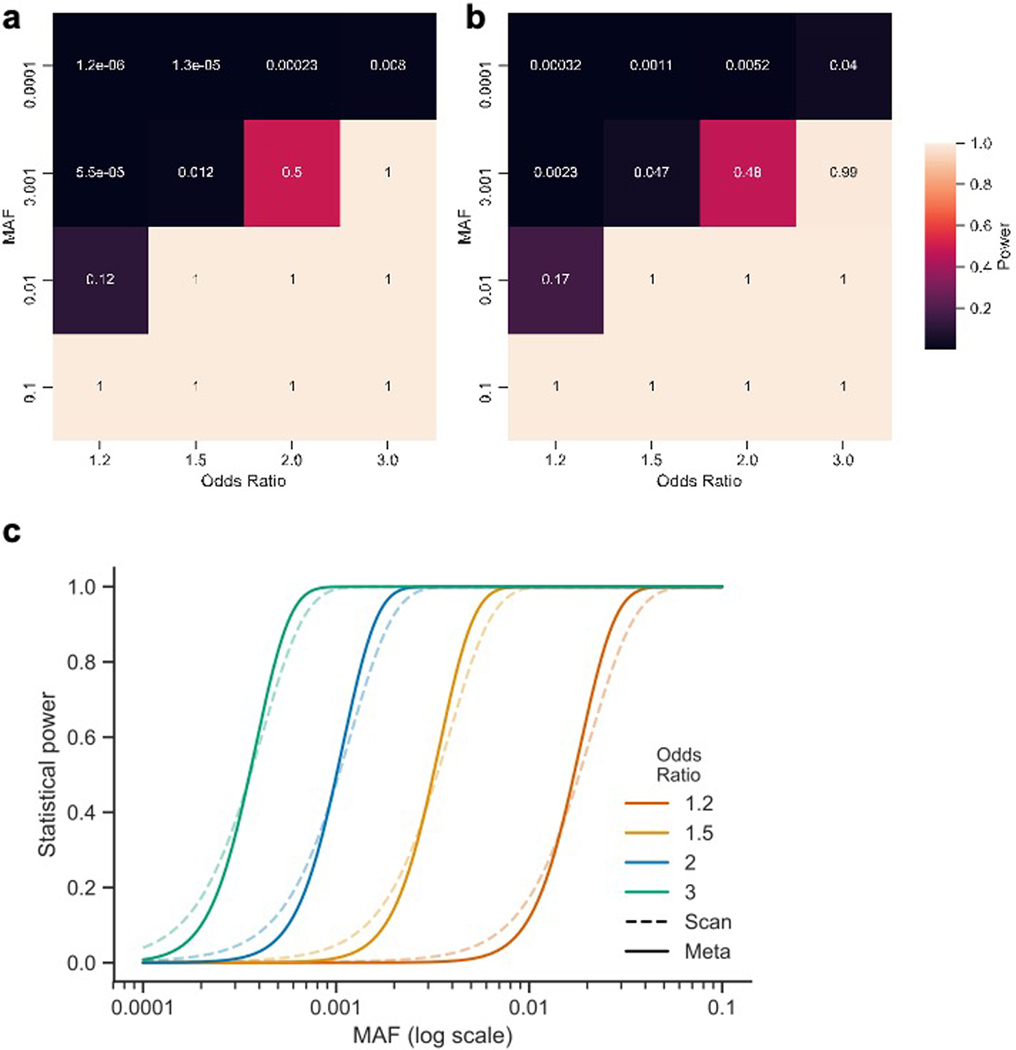
Extended Data Fig. 4. Power to detect single variant associations.
We performed a series of power calculations using the methodology described by Johnson and Abecasis (2017). Our initial ‘exome-wide scan’ (two cohorts) had fewer samples and a more lenient significance threshold than subsequent meta-analysis (five cohorts). However, both analyses had similar power to detect true associations at their respective significance levels. Our single-variant association analyses did not have the power to uncover association to variants with a MAF = 0.0001 and below (unless the variant has a very strong effect, e.g. 0.76 power at OR = 8). Similarly, the exome-wide scan had limited power to detect association to variants with a MAF = 0.001 and OR < 2, but was well-powered above these thresholds. a, Power of the exome-wide scan analysis b, Power of the meta-analysis. c, Power to detect single-variant associations at different minor allele frequencies at α = 0.0002 (‘scan’; dashed lines) and 3 x 10–7 (‘meta’; solid lines) and assuming Crohn’s disease population prevalence of 276 in 100,000, and an additive effect model.