Extended Data Fig. 9. Alterations in rarely mutated genes affecting gene expression.
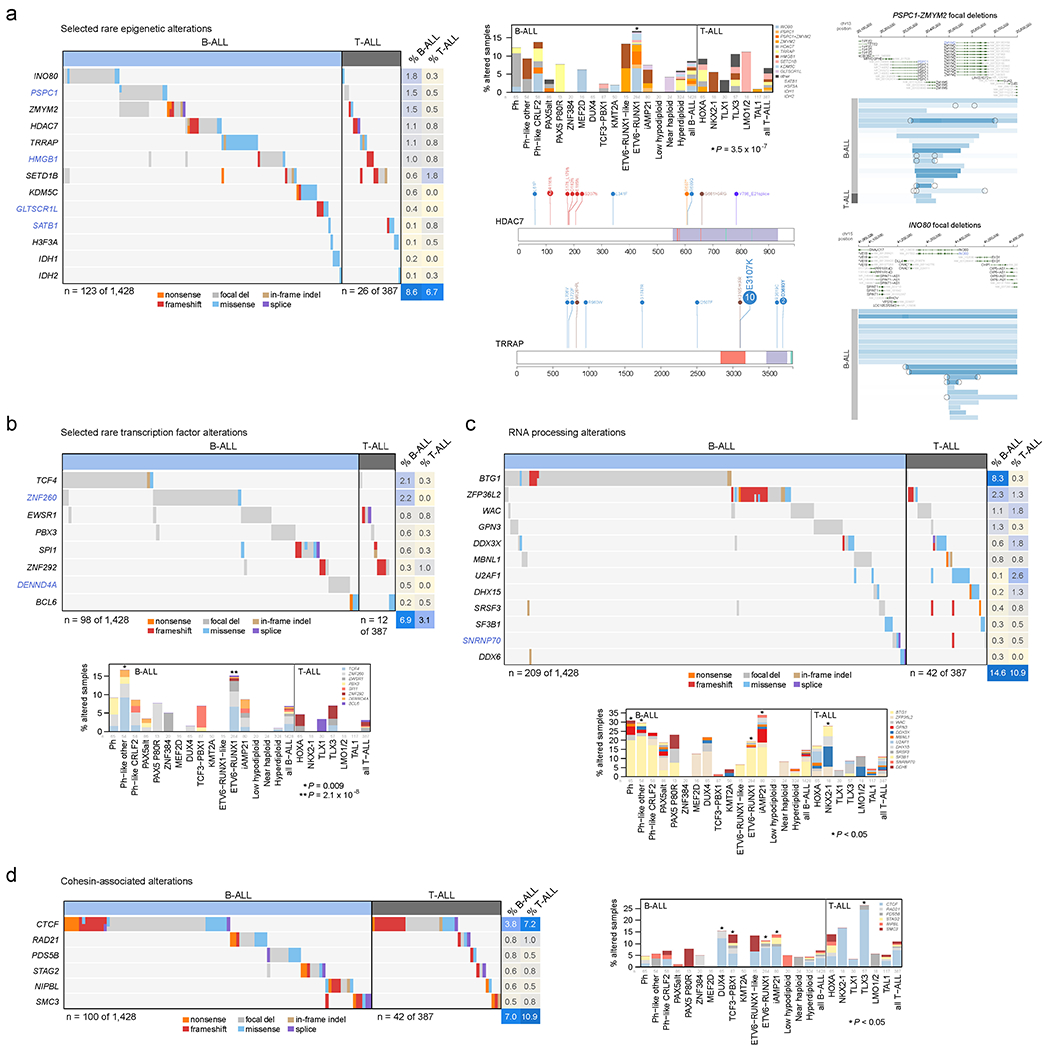
(a) Selected alterations in rare epigenetic modulators. Putative cancer driver genes are shown in blue text. Left shows an oncoprint showing only samples with alterations in at least one of these genes, with alteration type indicated by color and the percentage of samples in B-ALL or T-ALL altered at right. Top-middle shows the percentage of each subtype with alterations in these genes, color-coded by the specific gene altered. In samples with alterations with more than one gene, only the top-most gene in the legend is shown. Number of samples in each subtype is as in Fig. 5b. Right shows example gene alterations, including focal deletions (5 Mb or less; blue indicates degree of copy loss in each sample (row) and circles indicate SVs which were available for WGS samples only) in PSPC1-ZMYM2 and INO80. Sites of sequence alterations in HDAC7 and TRRAP are shown at middle-bottom. (b-d) Oncoprints and subtype bar plots as in (a) except that shown are selected transcription factors (b), RNA processing factors (c), and cohesion-associated genes (d). P values (asterisks) are by two-sided Fisher’s exact test comparing prevalence in the indicated subtype vs. all samples not belonging to that subtype (within that lineage (B-ALL or T-ALL), so that ETV6-RUNX1 subtype would be compared to B-ALL samples of other subtypes, while TLX3 subtype would be compared to T-ALL samples of other subtypes). In (c), exact P values are 8.6 x 10−4 (Ph), 0.0047 (Ph-like other), 0.015 (ETV6-RUNX1), 9.7 x 10−6 (iAMP21), and 0.035 (NKX2-1). In (d), exact P values are 0.020 (DUX4), 0.016 (TCF3-PBX1), 0.0027 (ETV6-RUNX1), 0.023 (iAMP21), and 2.6 x 10−4 (TLX3).
