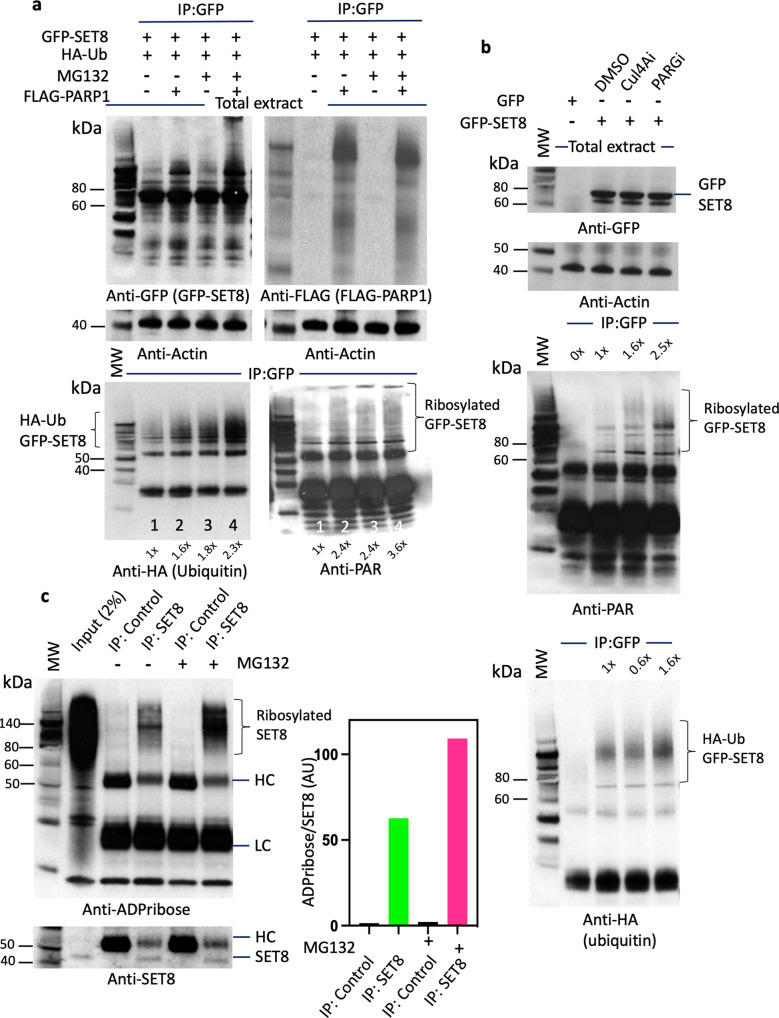
Fig. 3. PARP1 promotes SET8 protein degradation.
a GFP-SET8 immunoprecipitation in overexpressed GFP-SET8 COS-7 cells with or without FLAG-PARP1 overexpression in presence or not of the proteasome inhibitor MG132. Western blots detecting the amount of GFP-SET8 protein overexpressed in total extract (top, left) as well as the amount of FLAG-PARP1 protein overexpressed (top, right) using anti-GFP and anti-FLAG antibody, respectively. Western blots detecting the amount of ubiquitin (Ub) (bottom, left) or ADP-ribosylation (bottom, right) of immunoprecipitated GFP-SET8 protein using anti-HA and anti-ADP ribose antibody, respectively. Anti-actin was used as a loading control (middle). The fold increase was calculated by densitometry and indicated at the bottom of the western blot. b GFP-SET8 immunoprecipitation in overexpressed GFP-SET8 COS-7 cells in presence or not (DMSO) of Cullin inhibitor (Cul4Ai) or PARG inhibitor (PARGi). Western blot detection of total GFP-SET8 protein levels overexpressed in COS-7 cells (1st panel). Anti-actin antibody was used as control (2nd panel). Western blot detecting the amount of poly-ADP ribosylation, using anti-PAR antibody (3rd panel) or the amounts of HA-ubiquitin using anti-HA antibody (4th panel) of immunoprecipitated GFP-SET8 protein. c Endogenous SET8 immunoprecipitation in HeLa cells in presence or absence of the proteasome inhibitor MG132. Western blot (left side) detecting the amount of ADP-ribosylation in GFP (IP: control), SET8 immunoprecipitates (top panel) as well as the amount of SET8 protein (bottom panel). Respective densitometry analyses of SET8 ADP-ribosylation abundance representative of at least 2 biological experiments are shown (right side; n = 2).