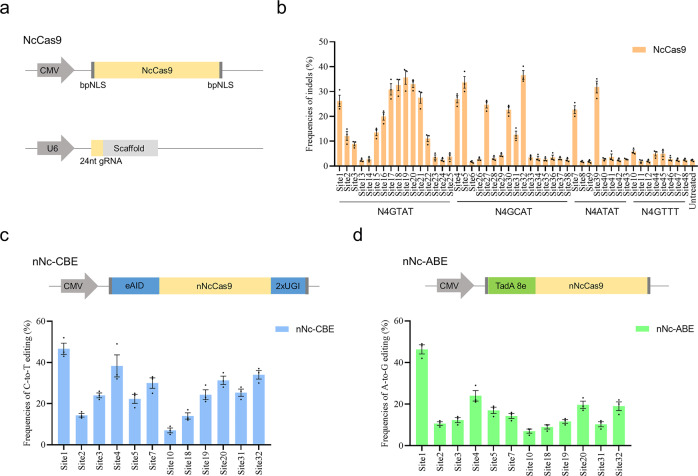
Fig. 3. Genome editing of NcCas9 and its base editors in human cells.
a Schematic of optimized NcCas9 and its sgRNA architecture. b Genome editing of NcCas9 with N4GYAT, N4ATAT and N4GTTT PAMs in 48 endogenous loci. c Top: Schematic of the nNc-CBE. Bottom: Efficient C-to-T conversions in 12 endogenous loci by four nNc-CBE. d Top: Schematic of the nNc-ABE, TadA 8e was used. Bottom: Efficient A-to-G conversions in 12 endogenous loci by nNc-ABE. The highest C-to-T/A-to-G editing frequency within the editing window represents the base editing frequency at each site. Error bars indicate s.e.m. (n = 3).