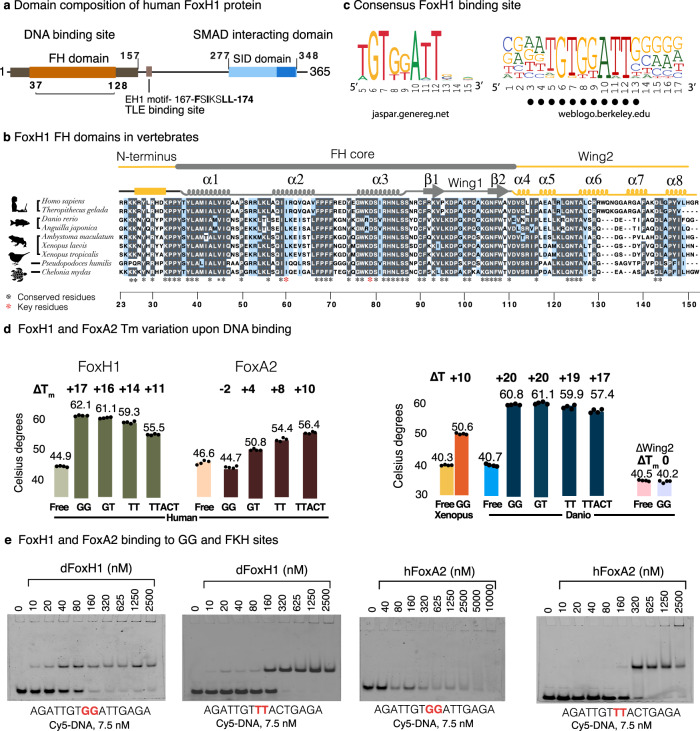
Fig. 1. The FoxH1 FH domain binds DNA with high affinity.
a Domain composition of FoxH1 proteins. The FH and SID domains are conserved in vertebrates. b Sequence alignment of FoxH1 FH domain in vertebrates. Secondary structure elements observed in the human complex are depicted on the top of the alignment. The FH core domain and the extended N- and C-termini are also indicated. Alignments were generated with MAFFT76 and the BoxShade server (https://bio.tools/boxshade). Residue conservation is color coded: white: low similarity, blue: high similarity, grey: strictly conserved. c Consensus DNA binding. The sequences used to derive this consensus are shown in Supplementary Fig. 1b. Base pairs found to participate in specific protein contacts are indicated. This consensus is almost identical to the JASPAR profile https://jaspar.genereg.net/. d Melting temperature modification of the human FoxH1 and FoxA2 domains in the presence of the different DNAs. The stability of the construct without the C-terminal extension (∆Wing2) is not affected by the presence of DNA. FoxA2 incubation with the GG site induced a decrease in its melting temperature. Melting temperatures correspond to two repetitions and three replicates. Values are summarized in Supplementary Table 1. Source Data are provided as a source data file. e Comparison of the binding properties of the FoxH1 and FoxA2 FH domains and two 16 bp cy5-labeled DNA molecules followed by native electrophoretic mobility shift assay (EMSA). The GG motif (in red) is derived from the native Gsc sequence. The TT sequence (in red) corresponds to a forkhead site. Whereas FoxH1 and FoxA2 bind well to the forkhead site, binding to the GG site is observed only for FoxH1.