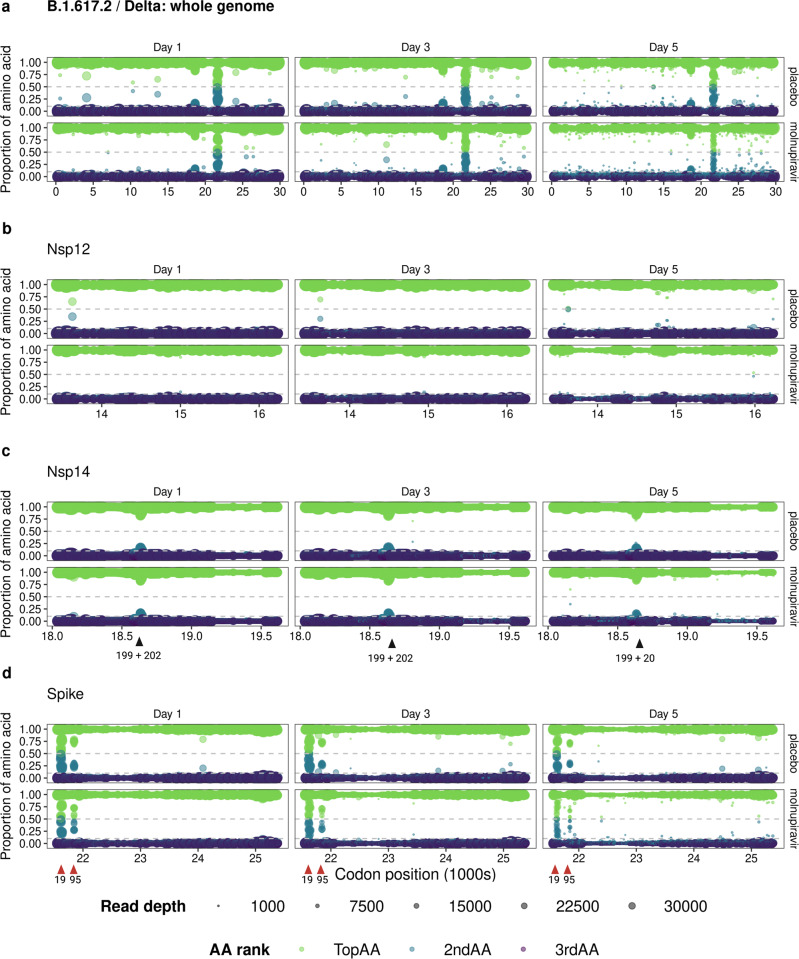
Fig. 2. Predicted amino acid variations derived from SARS-CoV-2 RNA in the whole genome, NSP12, NSP14 and Spike sequences.
a Predicted amino acid variation derived from RNA sequence information across the whole genome in all Delta infected participants (n = 52). Each sample is assigned a predicted “Top” (green), “2nd” (blue) and “3rd” (dark purple) amino acid (AA) based on proportion of reads at every genome position. Minimum read depth = 200. Minor genomic variants (>0.1 and <0.5; grey dashed lines) increase in frequency over time, with viral RNA from molnupiravir treated participants showing more diversity. b NSP12 showed very little minor genomic variation over the five days. c NSP14 also showed minor genomic stability, but had sites of low-level minor variation at codon positions 18,634 and 18,643 (indicated as amino acids 199 and 202 with black arrows) that were present in all samples tested and may represent a persistent sub-population. d Spike had two sites with an amino acid mixed population at codon positions 21,617 and 21,845 (indicated as amino acids 19 and 95 with red arrows) in all Delta samples analysed. These are known VOC sites in all the Delta sub-lineages.