Figure 1.
Transmission electron microscopy and tomography workflow
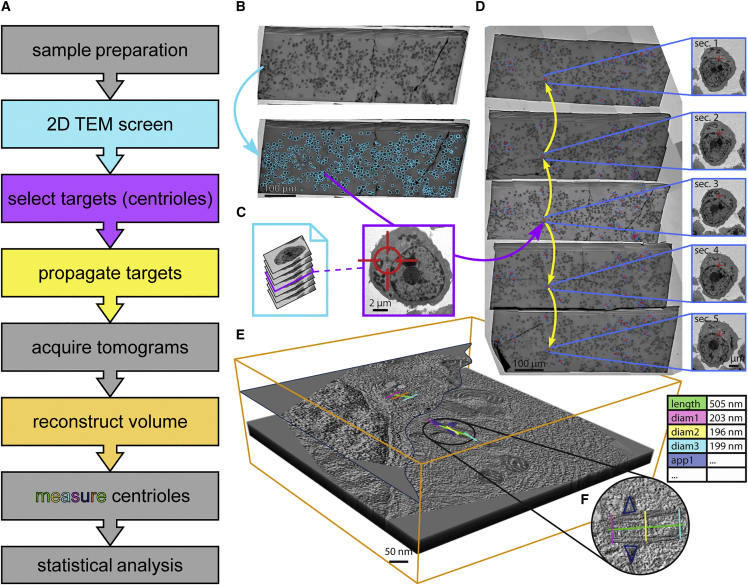
(A) Overview of all relevant aspects of the workflow. Colors of the text boxes are matched with the respective procedure in (B)–(E). Steps not explicitly depicted are colored in gray.
(B) After sample preparation, a 2D overview of the central section is obtained at 400× magnification (326.9 nm/px), and cells are semi-automatically detected and labeled for acquisition (blue arrow).
(C) Labeled cells are acquired at a magnification suitable for manual screening (3,000×, 42.75 nm/px, in this study). Output is generated as an image stack (left) that allows for identification of cells containing subcellular target features (here: centrioles). An exemplary image of a plasma cell containing a centriole (red crosshair) at 3,000× magnification is depicted on the right.
(D) Selected cells (blue boxes) containing target centrioles (red crosses) on the central section are semi-automatically propagated and identified on all adjacent sections (yellow arrows). As an example, inlays on the right show the same cell containing target features (here: centrioles) on all five acquired neighboring sections. Subsequently, single-axis electron tomography (15,500× magnification, 1.55 nm/px; tilt range: −60° to 60°; increment: 1°) is performed at all targets on all sections.
(E) Corresponding tomograms from individual sections are reconstructed and joined to produce a final volume of at least 3.1 × 3.1 × 1 μm per target (X × Y × Z; orange bounding box).
(F) Target centrioles can be measured and morphometrically analyzed in these volumes. Statistical analysis of centriole parameters can eventually be performed using the exported measured parameters (exemplarily visualized as a table on the right). The volume shown in this panel can be interactively viewed in MoBIE as MMRR_06_Grid1_c442. For instructions, refer to the STAR Methods section of this paper. TEM, transmission electron microscopy.