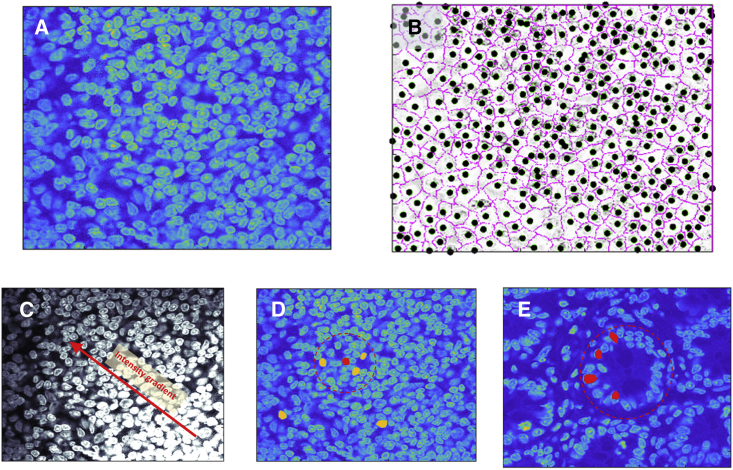
Figure 1.
Considering local and global spatial statistics and the influence of the tissue environment
(A and B) Computerized image analysis now readily provides segmentation of cell nucleus and cytoplasm in tissue and the location of the centroid position. In this example, (A) is a typical field of cells for which (B) shows the cell outlines (magenta line) and centroid positions (black circle) obtained by segmentation based on fluorescent staining of membrane and nucleus.
(C and D) Within quasi-homogeneous tissue, statistical analysis can be applied in a straightforward manner to identify spatial correlations of cell phenotypes or spatial patterning in cell markers. For example, in (C), a spatial gradient in pixel intensity is shown, such as would result from differing expression of fluorescent molecular biomarkers; while (D) depicts the definition of a local neighborhood (red dashed circle) within which the numbers of cells of specific types (shown in yellow and red) may be assessed for comparison to a hypothesis of random distribution.
(E) However, many biological samples will display a heterogeneous cellular environment, and in this case, care must be taken. Here, the image shows a tissue cross section with a clear meta-structure in which the cell positions are primarily determined by the tissue morphology. Now the definition of a cell neighborhood will lead to a test statistic for the selected cells (highlighted in red) that is highly significant, i.e., indicative of a pattern that is far from random. We need to be aware that, while we have a significant statistic, we do not have a significant result; this is just a reflection of the underlying tissue morphology rather than a directed biological response.