Figure 2.
Extracting spatial metrics from tissue images
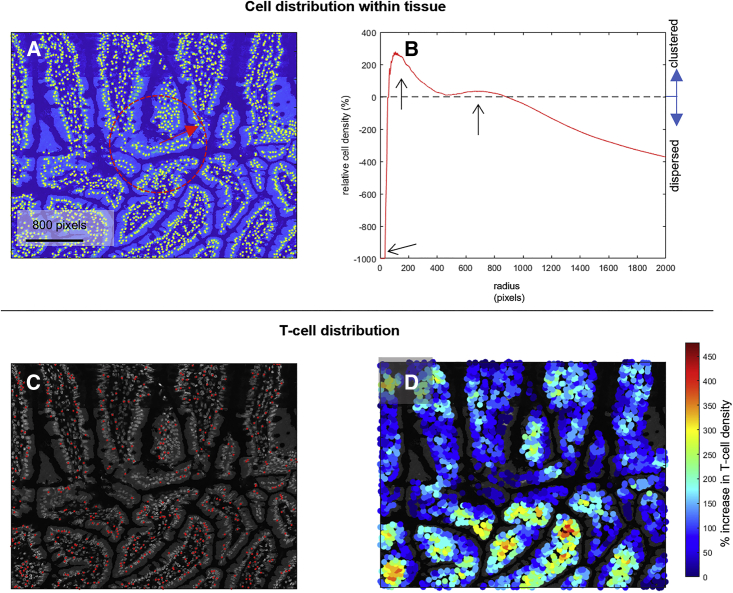
(A) Highly informative statistics can be extracted from straightforward calculation of spatial cell density. The image shows a section of mouse ileum where the centroids of all cells are highlighted in yellow. These can be used to calculate cell density within a given region, in this case, the circular region indicated in red.
(B) The result for image (A) is plotted in (B) as the relative percentage increase in area density compared with the random distribution value. Three features are immediately apparent in the plot (shown with arrows): (1) no cells within a radius of 30 pixels, as this is the minimum cell diameter; (2) a peak at ∼100 pixels corresponding to the typical cell separation within the large cell clusters; and (3) a second peak at ∼800 pixels corresponding to the mean separation of the clusters themselves, i.e., the spacing introduced by the presence of voids in the tissue.
(C) The same image from (A) can be analyzed to show only a specific sub-set of cells (T cells, identified by the presence of the CD3 cell differentiation marker and with centroid positions highlighted in red). Here, any deviation from random spatial patterning cannot be reliably ascertained by eye.
(D) When the image is re-plotted using a false color scale to display the local T cell density, this re-imaging clearly shows higher than expected T cell numbers within the lower half of the section.