Figure 3.
scRNA-seq characterization of iPSC-derived SNs
Cultures were processed for scRNA-seq on day 35. Sample data were analyzed separately before being integrated to create a normalized and controlled dataset.
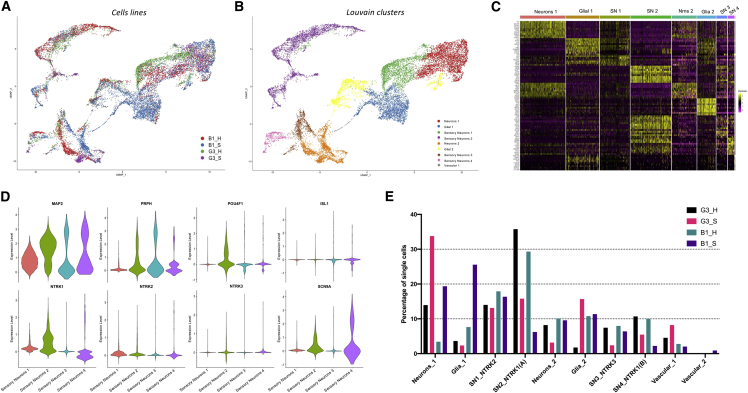
(A and B) UMAP dimension reduction plots of the combined dataset, created from principal-component analysis (PCA) of normalized gene expression. Cells were grouped by the original source sample from which they derived. (B) shows cells grouped by identified clusters, as determined by shared nearest neighbor-based clustering. A total of 10 clusters were identified.
(C) An expression heatmap highlighting the normalized expression of the top 10 marker genes identified for each of the 10 clusters. Clusters were assigned a cell type identity based on the differential expression of these markers.
(D) Violin plots presenting normalized gene expression of key genes for the cells from all conditions assigned in one of the SN clusters. The proportion of each of the total single cells for each of the conditions was determined based on their assignment to each of the 10 clusters.
(E) The percentage of each cell line that was clustered as part of each cell type.