Figure 5.
Investigating pathways associated with pain models using RNA-seq
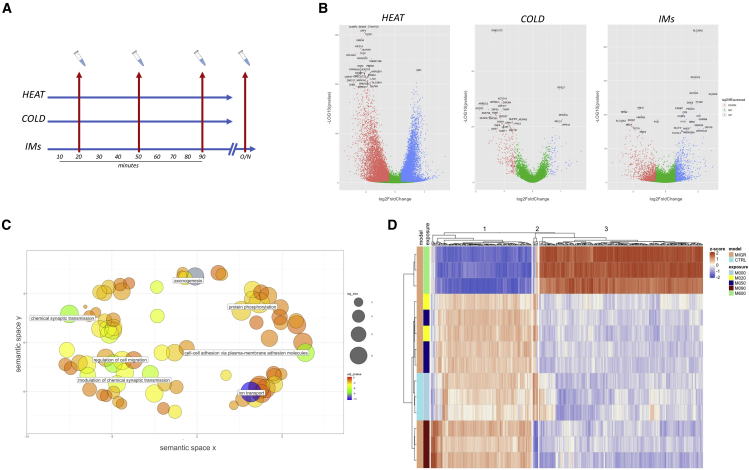
SNs were exposed to heat (45°C), cold (5°C), or an IM model over a total period of 18 h.
(A and B) Control samples were taken before stimulus exposure. DEGs were found for all models, with the volcano plots in (B) showing the log2 fold change (x axis) and –log10(p value) (y axis) for each DEG. Red and blue dots represent genes with FDR < 0.05 and a log2 fold change of ±0.6 (classed as significantly differentially expressed).
(C) The key enriched pathways for the inflammatory model as determined by analysis using Enrichr, followed by pathway condensation using Revigo. Each bubble shows an enriched pathway, positioned according to its functional relationship to other GO terms; labels denote representative pathways for each associated cluster. The size of bubbles shows the number of genes associated with each pathway term; the color scale shows the normalized adjusted p value of enrichment for each GO term.
(D) Heatmap of DEGs determined after exposure to IMs, with samples clustered with regard to exposure time. Normalized gene expression changes for each gene/sample are shown as Z scores.