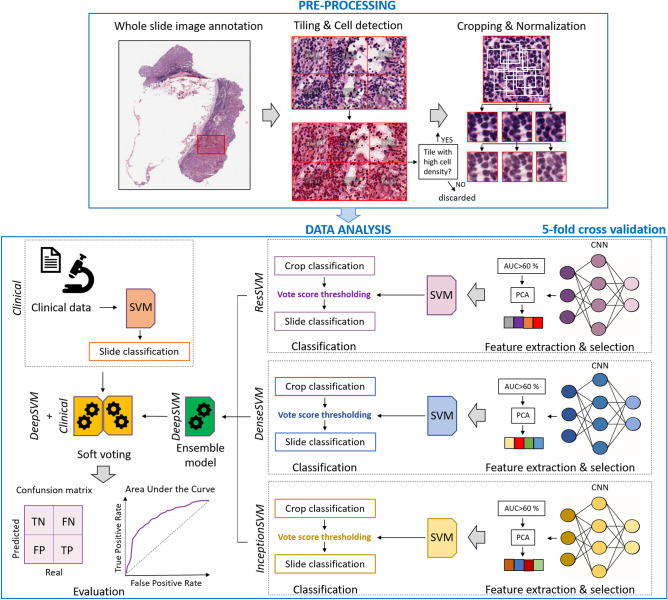
Figure 3.
Workflow of the proposed approach. Image pre-processing was firstly performed: WSIs were annotated by expert pathologists; the identified Region of Interests (ROIs), one per WSI, were tessellated into tiles of 224 × 224 pixels; cell detection was performed and only tiles with high cell density were retained and then divided into low-dimensional crops, which were finally colour-normalized. Data-analysis was then performed: crops were taken in input by three pre-trained CNNs. Each CNN extracted thousands of imaging features, later undergoing to a feature selection process; the selected features were employed to build a SVM classifier, which expressed a decision (DF or non-DF) firstly at crop level and then at slide level through the implementation of a vote score thresholding. In correspondence of the three CNNs, three models, called ResSVM, DenseSVM and InceptionSVM were defined. An ensemble model, named DeepSVM, was designed by combining the classification scores of the three models. A model, named Clinical, which took in input clinical data and used a SVM classifier to give in output classification scores at WSI level, was defined. A soft-voting procedure was implemented to combine the classification scores of DeepSVM and Clinical at WSI level and then at patient level. The final model was called DeepSVM + Clinical. Predictive performances were assessed by standard evaluation metrics.