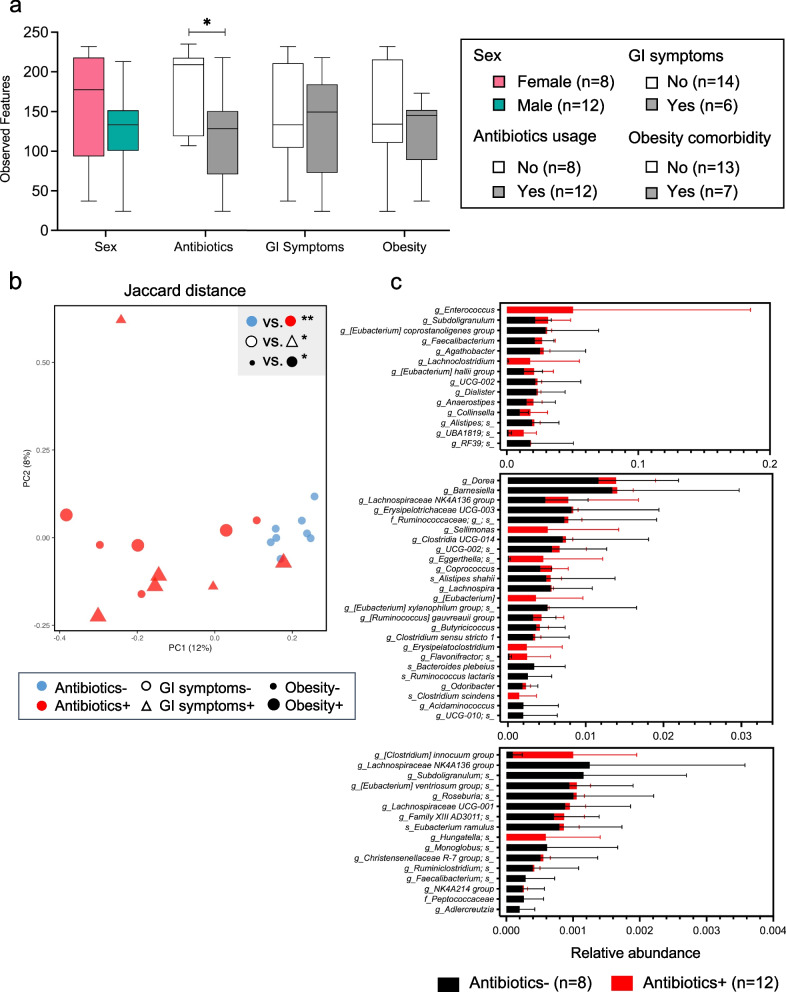
Fig. 5.
Gut microbial diversity and compositions in COVID-19-positive patients at sequence depth of 10,000. a Species richness was estimated by observed OTUs and evaluated based on subject sex, antibiotic use status, and the presence of GI symptoms, or of obesity comorbidity. Statistical significance was determined for comparisons between patients with or without antibiotic use by unpaired Student’s t-test: *p < 0.05. b Principal coordinates analysis (PCoA) was used for visualization of Jaccard dissimilarity of gut microbial communities for patients with or without antibiotic use, with or without GI symptoms, or with or without the comorbidity of obesity. The key in Fig. 5a indicates the number of subjects in each group. *p < 0.05 and **p < 0.01 by PERMANOVA. c Significantly differential bacterial species in the 20 COVID-19-positive patients, organized according to antibiotic use, were identified by MaAsLin2 and stratified into groups of high- (top panel), medium- (middle panel), and low- (bottom panel) relative abundances