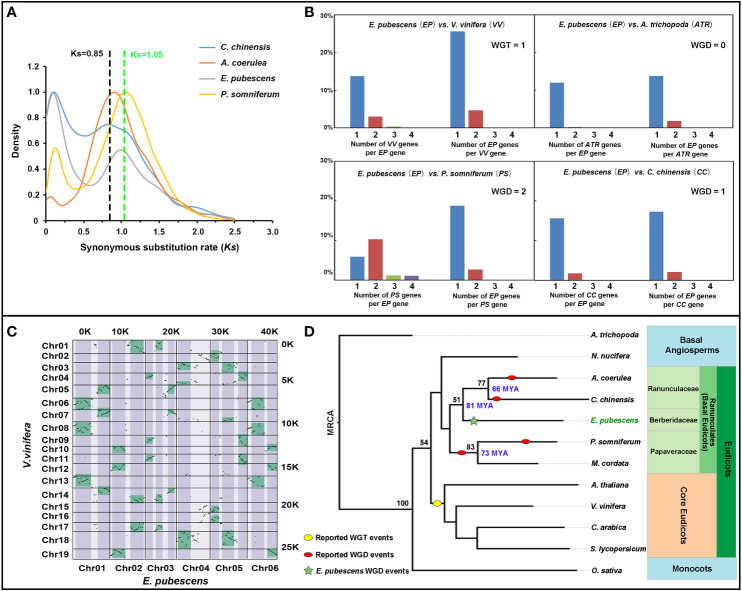
Figure 3.
Comparative genomic analysis of E. pubescens and other species. (A) Ks plots of orthologous genes in the genome of C. chinensis, A. coerulea, E. pubescens, and P. somniferum. Blue, red, light purple and yellow curves represent C. chinensis, A. coerulea, E. pubescens and P. somniferum respectively. (B) Comparison of orthologous gene ratio of E. pubescens to Vitis vinifera, P. somniferum, Amborella trichopoda, and C. chinensis, reciprocally. Blue, red, green and purple bars represent one, two, three and four orthologous genes. Short names were assigned as EP (E. pubescens), VV (V. vinifera), ATR (A. trichopoda), PP (P. somniferum) and CC (C. chinensis). Top left is the comparison of orthologous gene ratio of EP to VV; top right, the comparison of orthologous gene ratio of EP to ATR; bottom left, the comparison of orthologous gene ratio of EP to PP; bottom right, the comparison of orthologous gene ratio of EP to CC. (C) Dotplot of syntenic blocks between E. pubescens and V. vinifera. Chromosomes of V. vinifera and E. pubescens were labeled on the Y-axis and X-axis respectively; the number of syntenic blocks from V. vinifera and E. pubescens was labeled on the right and top respectively; syntenic blocks with 3:1 ratio of V. vinifera to E. pubescens were green-colored on the dotplot. (D) Phylogenetic tree of 12 plant species. In phylogenetic tree, red and yellow ovals represent the reported WGD events and WGT-γ event, respectively; the green star highlighted the independent WGD event in E. pubescens; ML (Maximum Likelihood Method) bootstrap values were labeled on the left of key phylogenetic tree nodes in black and the divergence times were labeled on the right of key phylogenetic tree nodes in blue, based on Timetree online services. Based on APG IV system, the phylogenetic classification of 12 plant species was labeled on the right of phylogenetic tree with different background color.