Abstract
Background & Aims
During liver regeneration after partial hepatectomy, the function and metabolic pathways governing transient lipid droplet accumulation in hepatocytes remain obscure. Mammalian target of rapamycin 2 (mTORC2) facilitates de novo synthesis of hepatic lipids. Under normal conditions and in tumorigenesis, decreased levels of triglyceride (TG) and fatty acids (FAs) are observed in the mTORC2-deficient liver. However, during liver regeneration, their levels increase in the absence of mTORC2.
Methods
Rictor liver-specific knockout and control mice underwent partial hepatectomy, followed by measurement of TG and FA contents during liver regeneration. FA metabolism was evaluated by analyzing the expression of FA metabolism-related genes and proteins. Intraperitoneal injection of the peroxisome proliferator-activated receptor α (PPAR-α) agonist, p53 inhibitor, and protein kinase B (AKT) activator was performed to verify the regulatory pathways involved. Lipid mass spectrometry was performed to identify the potential PPAR-α activators.
Results
The expression of FA metabolism-related genes and proteins suggested that FAs are mainly transported into hepatocytes during liver regeneration. The PPAR-α pathway is down-regulated significantly in the mTORC2-deficient liver, resulting in the accumulation of TGs. The PPAR-α agonist WY-14643 rescued deficient liver regeneration and survival in mTORC2-deficient mice. Furthermore, lipidomic analysis suggested that mTORC2 deficiency substantially reduced glucosylceramide (GluCer) content. GluCer activated PPAR-α. GluCer treatment in vivo restored the regenerative ability and survival rates in the mTORC2-deficient group.
Conclusions
Our data suggest that FAs are mainly transported into hepatocytes during liver regeneration, and their metabolism is facilitated by mTORC2 through the GluCer–PPAR-α pathway, thereby establishing a novel role for mTORC2 in lipid metabolism.
Keywords: Fatty Acids, Hepatectomy, Metabolism, Proliferation, Triglyceride
Abbreviations used in this paper: AKT, protein kinase B; BrdU, 5-Bromo-2'-deoxyuridine; CD36, cluster of differentiation 36; FA, fatty acid; GluCer, glucosylceramide; KO, knockout; LW/BW, liver-to–body weight ratio; mRNA, messenger RNA; MS, mass spectrometry; mTORC2, mammalian target of rapamycin complex 2; PCNA, proliferating cell nuclear antigen; PFT-α, pifithrin-α; PH, partial hepatectomy; PPAR-α, peroxisome proliferator-activated receptor α; Rictor, RPTOR-independent companion of mammalian target of rapamycin complex 2; R-LKO, liver-specific knockout Rictor; SP, sphingolipid; SPT, serine palmitoyltransferase; VLFA, very long-chain fatty acids; TG, triglyceride
Graphical abstract
Summary.
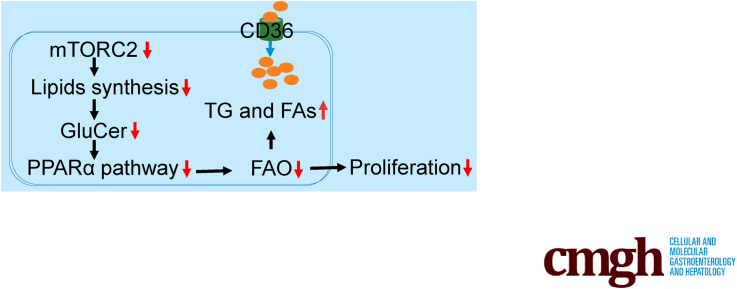
Fatty acids are mainly transported into hepatocytes during liver regeneration. Mammalian target of rapamycin complex 2 facilitates fatty acids oxidation through the glucosylceramide–peroxisome proliferator-activated receptor α pathway, thereby establishing its novel role in lipid metabolism.
The liver has a remarkable capacity for recovery from injury. Regenerative potential is essential for survival after liver damage caused by ischemia, viral infections, alcoholism, or drug overdose. However, inadequate liver regeneration remains a cause of morbidity and mortality induced by hepatic diseases, as well as tumor resection or living-donor liver transplantation.1, 2, 3 Elucidating the complicated mechanism of liver regeneration remains a crucial goal in hepatic research.4, 5, 6
In regeneration mouse models and human livers, fat accumulation occurs transiently and is required for physiological hepatic regeneration after partial hepatectomy (PH). Lipid droplets are formed mainly by triglyceride (TG) levels peaking at 24–48 hours after PH and reducing to baseline at 72 hours after PH. Both overload and deficiency of lipid droplets are harmful to hepatocyte proliferation during liver regeneration.7, 8, 9, 10, 11, 12 However, the mechanisms involved in lipid accumulation and lipid metabolism during liver regeneration remain obscure. Liver-specific knockout of the de novo lipid synthesis gene Fasn showed no change in 5-bromo-2′-deoxyuridine (BrdU)-positive hepatocytes.13 After PH, there was no time-dependent impairment in liver regeneration in PPAR-α knockout mouse models.13 Systemic knockdown of PPAR-α resulted in a 12- to 24-hour lag to the G1/S checkpoint.14 Liver-specific deletion of PPAR-α significantly reduced hepatocyte proliferation at 32 hours.15 The fatty acid (FA) transporter cluster of differentiation 36 (CD36) contributes to enhanced transient regeneration-associated steatosis.16 However, the accumulation and utilization of lipid droplets during liver regeneration and the physiological significance of this process remain unclear.
Mammalian target of rapamycin 2 (mTORC2) is a master regulator of lipid synthesis and regulatory associated protein Of MTOR Complex 1 (RPTOR)-independent companion of mTOR complex 2 (Rictor) is a key component of the mTORC2 complex.17 Liver-specific knockout Rictor (R-LKO) mouse models show liver-specific deficiency of mTORC2 and marked hyperglycemia, hyperinsulinemia, and hypolipidemia.18 Decreased lipid levels are reported in the mTORC2 knockout (KO) liver because of a remarkable deficiency in de novo lipid synthesis. The transcription of de novo lipid synthesis–related genes, such as Fasn, Acly, and Srebp1c, is inhibited significantly under normal conditions in R-LKO mice. mTORC2 also promotes hepatic tumorigenesis through lipid synthesis, with sphingolipids and cardiolipin showing a causal effect during this process.19 Prior studies have shown that mTORC2 is necessary for timely liver regeneration.20
In this study, both male and female R-LKO mice showed accumulated lipid droplets in the regenerated liver. Further data suggest the FAs are mainly transported into hepatocytes. mTORC2 deficiency leads to significantly down-regulated peroxisome proliferator-activated receptor α (PPAR-α) levels and excess lipid droplet accumulation. The PPAR-α pathway down-regulation causes impaired hepatocyte proliferation and poor survival in R-LKO mice. Furthermore, we identified glucosylceramide (GluCer) as a novel activator of the PPAR-α pathway, which activated the pathway in vivo, promoted hepatocyte proliferation, and enhanced the survival rate in R-LKO mice after PH. Therefore, mTORC2 promoted liver regeneration via the GluCer–PPAR-α pathway, thereby establishing its newly discovered role in FA oxidation.
Results
mTORC2 Deficiency Resulted in Decreased Hepatocyte Proliferation During Liver Regeneration After PH
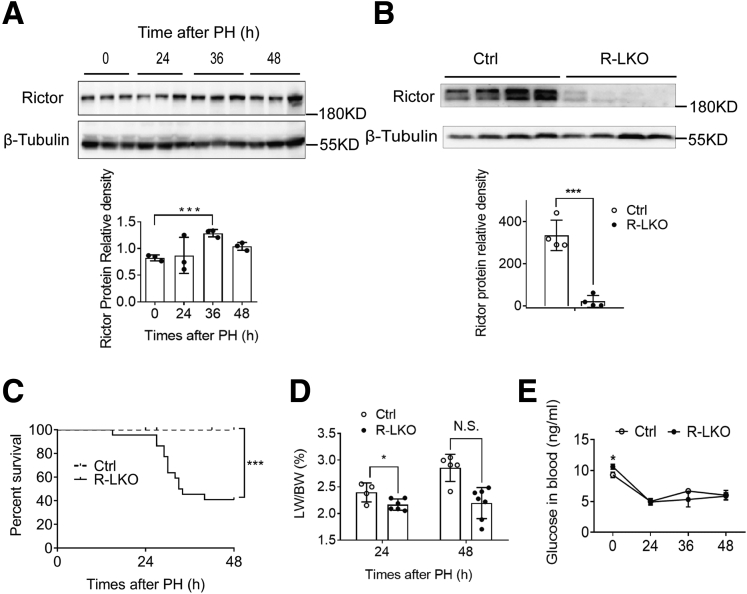
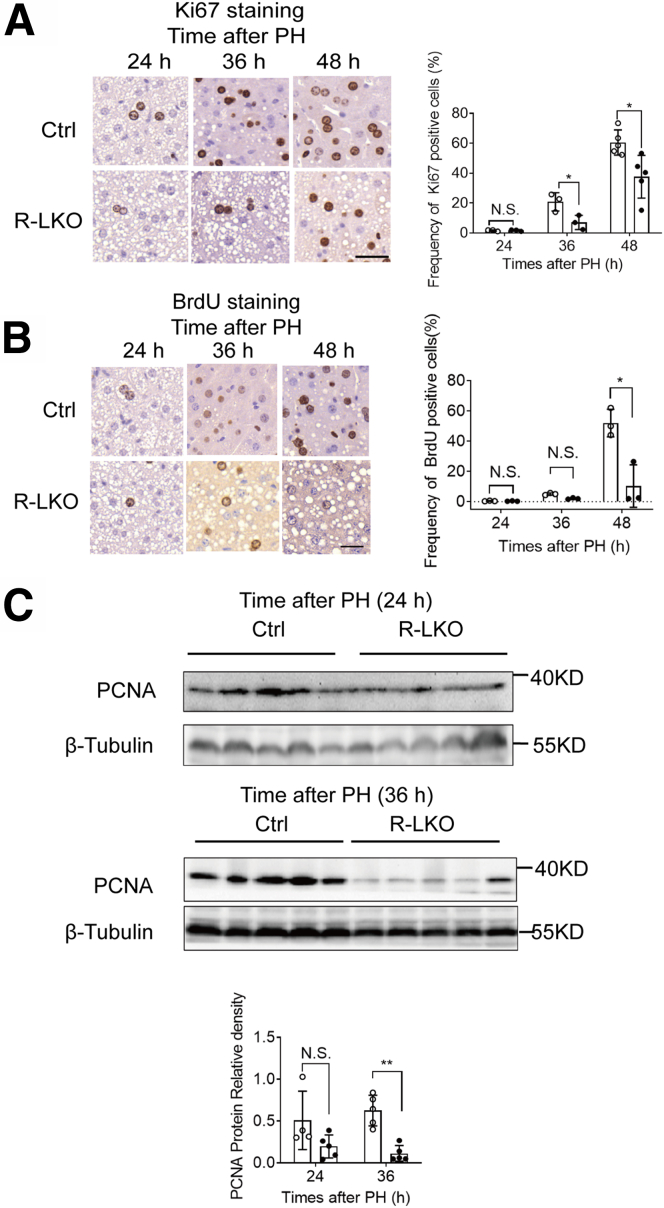
Rictor is a key component of the mTORC2 complex. To investigate the role of mTORC2 in liver regeneration, we examined Rictor expression in the liver after PH in wild-type mice. Rictor expression increased by 55% in the regenerated liver at 36 hours after PH (Figure 1A). To investigate the role of mTORC2 in liver regeneration, R-LKO mice were used (Figure 1B); both male and female mice were used unbiasedly. At 48 hours after PH, the survival rate of the R-LKO group was 41% (Figure 1C). The liver-to–body weight ratio (LW/BW) recovered in the control group; however, it did not increase in R-LKO mice, which survived until 48 hours after PH (Figure 1D). Blood glucose levels were higher (P < .05) in R-LKO mice before PH (Figure 1E). However, glucose levels decreased after PH in both the control and R-LKO groups provided access to food ad libitum, and there were no differences between the 2 groups (Figure 1E). The hepatocyte proliferation rate was decreased significantly (P < .05) in R-LKO livers compared with the rate seen in the control group after PH, as indicated by BrdU and Ki-67 incorporation assays (Figure 2A and B). Consistently, the peak expression levels of proliferating cell nuclear antigen (PCNA) were significantly lower (P < .01 at 36 hours) in the R-LKO livers in comparison with the corresponding levels in the control group after PH (Figure 2C). In summary, these results indicated that mTORC2 deficiency severely hampers hepatocyte proliferation after PH and mTORC2 is essential for liver regeneration.
Figure 1.
Decreased LW/BW and survival rate in mTORC2 deficient mice after PH. (A) Representative western blot analysis of rictor proteins in control mice liver samples after PH. Protein quantification was normalized to β-tubulin (n = 3 per group). (B) Representative Western blot analysis of Rictor proteins in liver samples isolated from control (Ctrl) and R-LKO mice. Quantifications were normalized to β-tubulin (n = 4 mice for each time point). (C) Survival curves of Ctrl (n = 20 mice) and R-LKO (n = 22 mice) mice after 2/3 PH. (D) LW/BW of Ctrl (green) and R-LKO mice at 24 and 48 hours after PH (4–6 mice per group). (E) Analysis of serum glucose in Ctrl and R-LKO mice at the indicated time points after 2/3 PH (n = 4 mice for each time point). Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3D HISTECH, Ltd). Error bars represent the means ± SD. ∗P < .05, ∗∗∗P < .001.
Figure 2.
Decreased hepatocytes proliferation after PH in R-LKO mice. (A) Representative immunohistochemistry staining and quantification of Ki-67–positive cells in liver tissue of control (Ctrl) and R-LKO mice after 2/3 PH. The dark nucleus indicates BrdU-positive samples. Three sections per mouse were examined (n = 4 mice for each time point). Scale bar: 20 μm. (B) Representative immunohistochemistry staining and quantification of BrdU-positive cells in livers of Ctrl and R-LKO mice after 2/3 PH. The dark nucleus indicates BrdU-positive samples. Three sections per mouse were examined (n = 4 mice for each time point). Scale bar: 20 μm. (C) Western blot analysis of hepatic PCNA expression in Ctrl and R-LKO mice at the indicated times after PH. Quantifications were normalized to β-tubulin (n = 5 mice at each time point). Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3D HISTECH, Ltd). Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3D HISTECH, Ltd). The VersaDoc Imaging System (Bio-Rad) and software were used to visualize proteins. ImageJ was used to quantify the band densities. Error bars represent the means ± SD. ∗P < .05, ∗∗P < .01.
Lipid Droplet Accumulation Increased in the mTORC2-Deficient Liver During Liver Regeneration
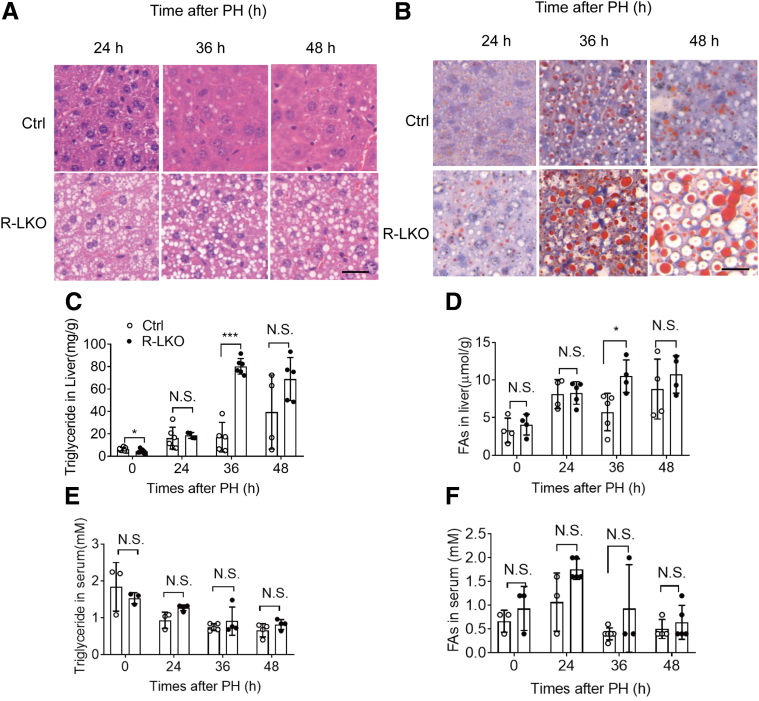
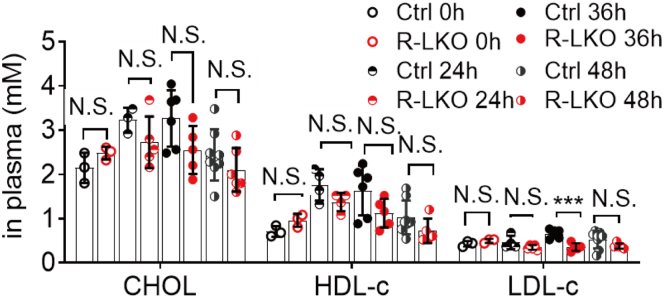
The R-LKO liver manifests hypolipidemia owing to decreased lipogenesis. However, more lipid droplets accumulated in the liver tissue of the R-LKO mice than in the liver tissue of the mice in the control group, as indicated by H&E and Oil Red O staining (Figure 3A and B). Accordingly, the biochemical index showed that TG content in the R-LKO livers was increased significantly (P < .001) 36 hours after PH (Figure 3C). FA levels also increased significantly (P < .05) in the R-LKO mice at 36 hours compared with the FA levels in the control group (Figure 3D); in addition, the serum levels of FA and TG remained similar to the corresponding serum levels in the normal group (Figure 3E and F). Levels of cholesterol, high-density lipoprotein cholesterol, especially low-density lipoprotein cholesterol, were decreased in the mTORC2 KO regenerated livers at 36 hours time point. (P < .001) compared with the corresponding levels in the control group (Figure 4). These data indicate that more TG and FAs accumulate in R-LKO mice during liver regeneration.
Figure 3.
Lipid droplet accumulation increased in the mTORC2 KO liver during liver regeneration. (A) H&E staining from control (Ctrl) and R-LKO mice at the indicated time points after 2/3 PH. Scale bar: 20 μm. (B) Analysis of lipids by Oil Red O staining performed with frozen liver sections at the indicated time points after 2/3 PH. Scale bar: 20 μm. (C) Analysis of TG contents in liver tissue isolated from Ctrl and R-LKO mice. Means ± SD, 4–12 mice per time point. (D) Analysis of FA contents in liver tissue isolated from Ctrl and L-RKO mice. Means ± SD, 4–5 mice per time point. (E) Analysis of TG contents in serum from Ctrl and L-RKO mice, 3–4 mice per time point. (F) Analysis of FA contents in serum from Ctrl and L-RKO mice. Means ± SD, 3–6 mice per time point. H&E and Oil Red staining slices were scanned and captured by Pannoramic MIDI (3DHISTECH, Ltd). ∗P < .05, ∗∗∗P < .001.
Figure 4.
Decreased CHOL level in serum after PH in R-LKO mice. Analysis of CHOL, HDL-c, and LDL-c in serum from control (Ctrl) and R-LKO mice (4–8 mice for each time point). Data are presented as means ± SD. ∗∗∗P < .001. CHOL, cholesterol; HDL-c, high-density lipoprotein cholesterol; LDL-c, low-density lipoprotein cholesterol.
Up-regulated Expression of Genes and Proteins Associated With FA Transportation During Liver Regeneration
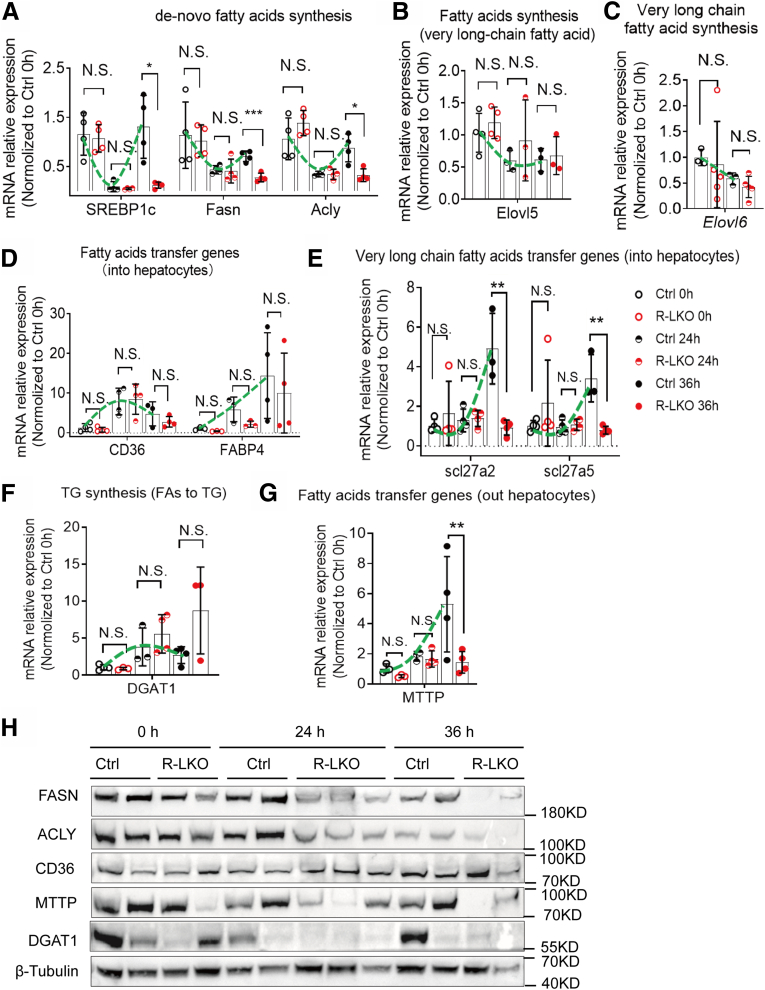
To elucidate the cause of lipid droplet accumulation during liver regeneration, we analyzed the mRNA expression of genes involved in FA metabolism in control mice. During liver regeneration, the mRNA expression levels of de novo FA synthesis genes, such as Srebp1c and Acly, were decreased significantly (P < .05) in the control livers at the 24-hour time point compared with the corresponding levels before PH (0 h) (Figure 5A, green line). The transcriptional levels of long-chain synthesis-relevant genes—Elovl5 and Elovl6—also were decreased after PH (Figure 5B and Figure 5C). These results indicate that de novo FA synthesis was inhibited in the first 24 hours after PH.
Figure 5.
The transportation of lipids was upregulated during liver regeneration. (A) Relative mRNA expression levels of de novo lipid synthesis genes in Ctrl (green line) or R-LKO mouse liver. The mRNA expression of sterol regulatory element-binding transcription factor 1c (SREBP1c), fatty acid synthase (Fasn), and adenosine triphosphate–citrate lyase; Acox1, acyl-CoA oxidase 1 (Acly) were normalized to the levels of control hepatocytes before PH (0 h). Means ± SD, 4 mice per time point. (B) Relative mRNA expression levels of very long chain fatty acid synthesis at the indicated time points in Ctrl (green line) or R-LKO mouse liver. The expression of Elvol5 is normalized to the levels of control hepatocytes before PH (0 h). Means ± SD, 3–4 mice per time point. (C) Relative mRNA expression levels of very long chain fatty acids transported into hepatocyte genes at the indicated time points in Ctrl (green line) or R-LKO liver. mRNA expression levels of slc27a2 and slc27a5 are normalized to the levels of control hepatocytes before PH (0 hours) (3–4 mice for each time point). (D) Relative mRNA expression levels of lipid transfer into hepatocytes at the indicated time points in Ctrl (green line) or R-LKO mouse liver. The expression of CD36 and FABP4 were normalized to the levels of control hepatocytes before PH (0 h). Means ± SD, 3–4 mice per time point. (E) Relative mRNA expression levels of triglyceride (TG) synthesis gene DGAT1 at the indicated time points in Ctrl (green line) or R-LKO liver. mRNA expression levels of DGAT1 are normalized to the levels of control hepatocytes before PH (0 hours) (3–4 mice for each time point). (E) Relative mRNA expression levels of lipids transported out of hepatocyte genes at the indicated time points in Ctrl (green line) or R-LKO liver. mRNA expression levels of microsomal triglyceride transfer protein (MTTP) are normalized to the levels of control hepatocytes before PH (0 hours) (3–4 mice for each time point). (F) Relative mRNA expression levels of hydroxyacyl-CoA dehydrogenase (Hadh) at the indicated time points in Ctrl (green line) or R-LKO liver. mRNA expression levels are normalized to the levels of control hepatocytes before PH (0 hours) (4 mice for each time point). (G) Relative mRNA expression levels of fatty acid β-oxidation genes in peroxisome at the indicated time points in Ctrl (green line) or R-LKO liver. mRNA expression levels of Acyl-CoA Oxidase 1 (Acox) and acetyl-CoA acyltransferase 1 (Acaa1) are normalized to the levels of control hepatocytes before PH (0 hours) (4 mice for each time point). (H) Western blot analysis of lipid synthesis (Fasn, ACLY), secretion (microsomal triglyceride transfer protein [MTTP]), re-esterification (diacylglycerol acyltransferase 1 [DGAT1]), and uptake (cluster of differentiation 36 [CD36]) before (0 h) and after PH.
The messenger RNA (mRNA) expression levels of genes responsible for FA import into hepatocytes, such as Cd36 (P < .01) and Fabp4 (P = .05), were increased significantly in the livers of control mice at the 24-hour time point compared with the corresponding pre-PH (0 h) levels (Figure 5D, green line). Expression of Scl27a2 and Scl27a5, responsible for the transport of very long-chain FAs (VLFAs) into hepatocytes, was activated in the control group at 36 hours after PH (Figure 5E, green line). The expression of re-esterification gene Dgat1 was up-regulated during liver regeneration (Figure 5F). These data indicate that the FAs are transported into hepatocytes and synthesized into TGs.
Furthermore, the expression level of genes responsible for the export of FAs from hepatocytes, such as Mttp,21 was increased 36 hours after PH compared with the level before PH (Figure 5G, green line). Protein expression corroborated with the transcriptional-level data (Figure 5H).
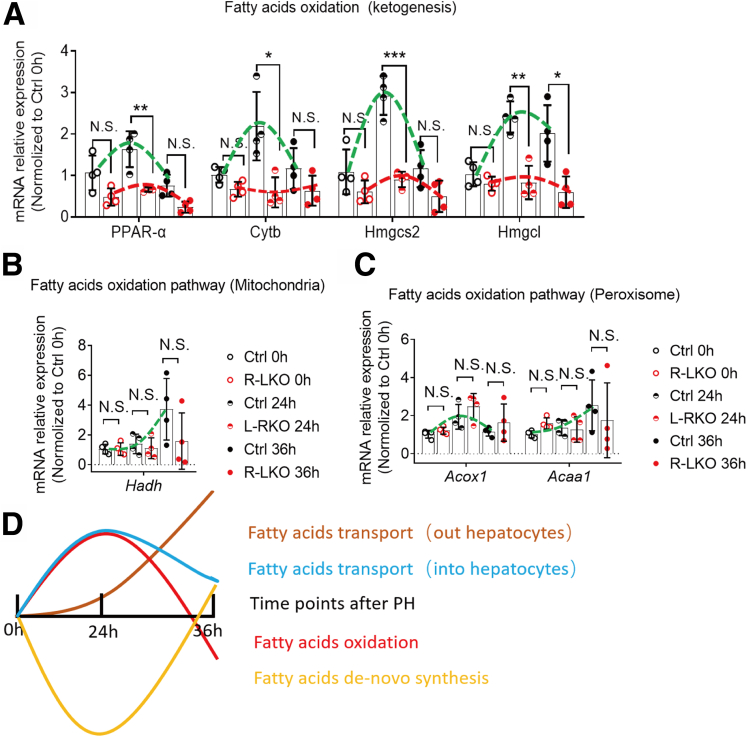
The expression of PPAR-α–mediated ketogenesis pathway genes, including Cytb, Hmgcs2, and Hmgcl, was activated 24 hours after PH in the control group (Figure 6A, green line). The mRNA expression of genes responsible for FA oxidation in the mitochondria, such as Hadh, also was activated 36 hours after PH (Figure 6B, green line). In addition, the expression of FA oxidation–associated genes in the peroxisome, such as Acox1 and Acaa1, which respond to VLFA oxidation, also was activated during liver regeneration (Figure 6C, green line).
Figure 6.
The PPAR-α pathway was decreased in mTORC2 KO liver during liver regeneration. (A) Relative mRNA expression levels of ketogenesis-associated genes at the indicated time points in Ctrl (green line) or R-LKO liver (red line). The expression of PPAR-α, Cytb, Hmgcs2, and Hmgcl were normalized to the levels of control hepatocytes before PH (0 h). Means ± SD, 4 mice per time point. (D) Fatty acid metabolic model in the normal liver between 0 and 36 hours after PH. The VersaDoc Imaging System (Bio-Rad) and software was used to visualize proteins. ImageJ was used to quantify the band densities. ∗P < .05, ∗∗P < .01 and ∗∗∗P < .001.
Compensatory changes in the mRNA expression of genes associated with FA metabolism in the control group after PH were concluded between the 0- and 36-hour time points (Figure 6D). These results indicate that the generation of FA metabolites during liver regeneration is well orchestrated. We hypothesized that the source of lipid droplet accumulation was increased in cell transport of FAs, and not de novo synthesis.
The PPAR-α Pathway Was Decreased in mTORC2 KO Liver During Liver Regeneration
It has been reported that under refeeding conditions, genes involved in lipid synthesis are down-regulated significantly in the livers of R-LKO mice fed a high-fat diet.18 Accordingly, during liver regeneration, although more FAs and TG accumulated in the R-LKO regenerated liver, the expression of FA de novo synthesis–related genes was down-regulated in the R-LKO group (Figure 5A and H). The trend for changes in the expression of VLFA synthesis–related genes Elovl5 and Elovl6 overlapped with that of controls (Figure 5B and Figure 5C).
The transcriptional changes in the expression of FA transport genes (into the hepatocytes), such as Cd36 and Fabp4, in R-LKO mice were similar to those in the control group (Figure 5D and D), while the transcriptional levels of genes of the fatty acid transport protein (FATP) family, such as Scl27a2 and Scl27a5, were decreased significantly in the R-LKO group at 36 hours (P < .01) (Figure 5E). These changes in gene expression may not be the sole cause of lipid accumulation in the R-LKO liver. β-FA oxidation in the mitochondria and peroxisomes produced no significant changes in comparison with the observations recorded in the control group (Figure 6B and C).
Activation of the ketogenesis-related PPAR-α pathway was decreased significantly in R-LKO livers (Figure 6A). Specifically, the mRNA expression levels of Pparα (P < .01) and its downstream genes Cytb (P < .05), Hmgcs2 (P < .001), and Hmgcl (P < .01) all were significantly lower than the corresponding levels in the control group, especially at the 24-hour time point (Figure 6A, green and red lines). Furthermore, the mRNA levels of genes responsible for transporting FAs out of hepatocytes, such as Mttp, also were decreased significantly in R-LKO mice at the 36-hour time point (P < .05) (Figure 5H and Figure 5G). Considering the time-dependent changes in microsomal triglyceride transfer protein expression after changes in PPAR-α pathway activity, we speculated that changes in microsomal triglyceride transfer protein are concomitant rather than causative.
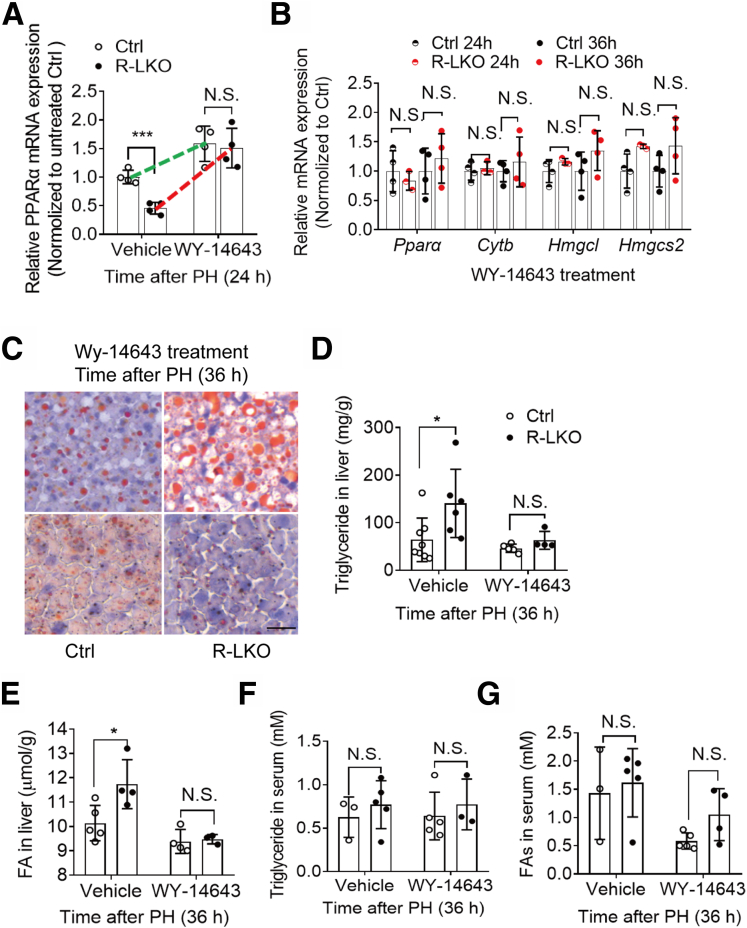
WY-14643 Ameliorated Lipid Accumulation in R-LKO Mice
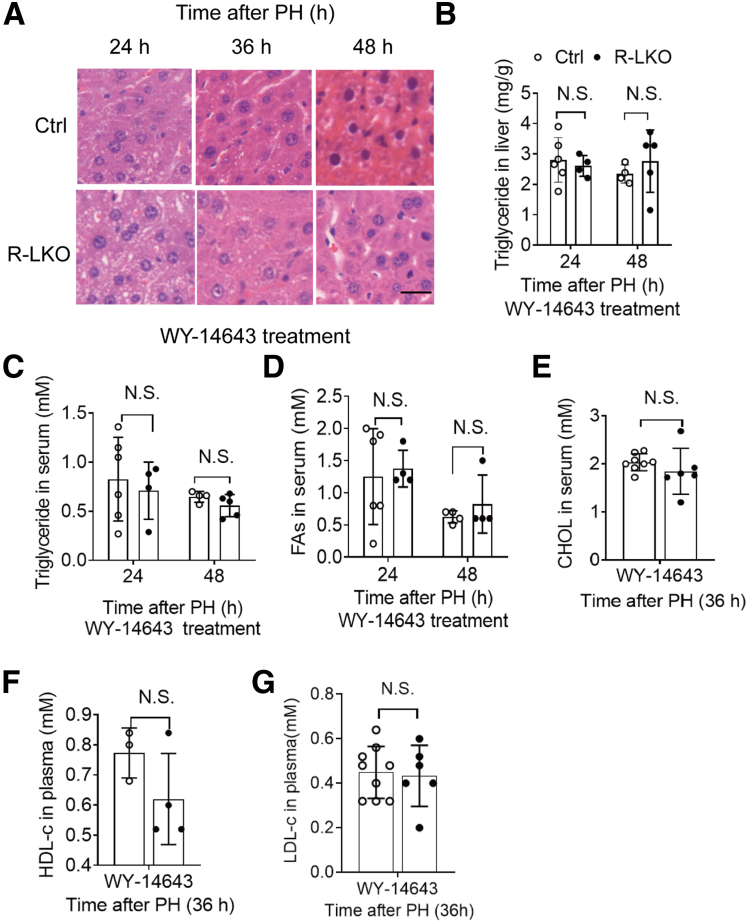
The transcriptional levels of lipid metabolism-related genes showed that inactivation of the PPAR-α pathway is responsible for the deposition of lipid droplets in the liver. To test whether lipid deposition is a factor governing deficient liver regeneration, we treated control and R-LKO mice with the PPAR-α agonist WY-14643.22 As anticipated, PPAR-α expression was increased after WY-14643 treatment in the livers of control and R-LKO mice (Figure 7A). The activation of the PPAR-α pathway in the R-LKO mouse livers increased to the same level as that in the control group livers after WY-14643 treatment (Figure 7B). Remarkably, in contrast to vehicle treatment, WY-14643 treatment completely abolished the differences in TG and FA contents in the hepatic tissues between control and R-LKO mice at 36 hours after PH and showed the same levels in the serum (Figure 7C–G and Figure 8A–D). Furthermore, total cholesterol levels, including high-density lipoprotein cholesterol and low-density lipoprotein cholesterol levels, increased, reaching the levels observed in the control group after WY-14643 treatment (Figure 8E–G).
Figure 7.
PPARα agonist equilibrates lipids contents during regeneration in mTORC2 absent liver. (A) mRNA relative expression of PPARα in WY-14643– or vehicle-treated mice 24 hours after PH. Green, control; red, R-LKO. Means ± SD, 4 mice per time point. ∗∗∗P < .001. (B) Relative mRNA expression levels of hepatic ketogenesis genes at the indicated time points after WY-14643 treatment. The expression of peroxisome proliferator-activated receptor alpha (PPARα), cytochrome b (Cytb), 3-Hydroxy-3-Methylglutaryl-CoA Synthase 2, (Hmgcs2), and 3-Hydroxy-3-Methylglutaryl-CoA Lyase (Hmgcl) were normalized to the levels of control hepatocytes before PH (0 hours) (3–4 mice for each time point). (C) Analysis of lipids by Oil Red O staining performed with frozen liver sections 36 hours after PH. Scale bar: 20 μm (right). (D) Hepatic content of TGs from vehicle and WY-14643–treated control (Ctrl) and R-LKO mice 36 hours after PH. Means ± SD, 4–8 mice per time point. ∗P < .05. (E) Hepatic content of FAs in liver tissue isolated from vehicle and WY-14643–treated Ctrl and R-LKO mice 36 hours after PH. Means ± SD, 3–5 mice per time point. ∗P < .05. (F) Analysis of TG contents in serum from vehicle and WY-14643–treated Ctrl and R-LKO mice 36 hours after PH. Means ± SD, 3–5 mice per time point. (G) Analysis of FA contents in serum from vehicle and WY-14643–treated Ctrl and R-LKO mice 36 hours after PH. Means ± SD, 3–5 mice per time point. Oil Red staining slices were scanned and captured by Pannoramic MIDI (3DHISTECH, Ltd).
Figure 8.
PPARα agonist equilibrates lipids contents during regeneration in mTORC2 absent liver. (A) Representative hematoxylin staining of hepatocytes in WY-14643–treated Ctrl and R-LKO mice liver at the indicated time points after PH. Scale bar: 20 μm. (B) Hepatic content of TGs from WY-14643–treated Ctrl and R-LKO mice at the indicated time points after PH (4–6 mice for each time point). (C) Analysis of TG contents in serum from WY-14643–treated Ctrl and R-LKO mice at the indicated time points after PH (4–6 mice for each time point). (D) Analysis of FA contents in serum from WY-14643–treated Ctrl and R-LKO mice at the indicated time points after PH (4–6 mice for each time point). (E–G) Analysis of cholesterol (CHOL), high density liptein cholesterol (HDLC), and low density liptein cholesterol (LDLC) in serum from WY-14643–treated Ctrl and R-LKO mice 36 hours after PH (3–9 mice for each time point). The hematoxylin and eosin stain (HE) slices were scanned and captured by Pannoramic MIDI (3D HISTECH, Ltd, budapest, Hungary).
WY-14643 Improved Liver Regeneration and Survival Rate in R-LKO Mice
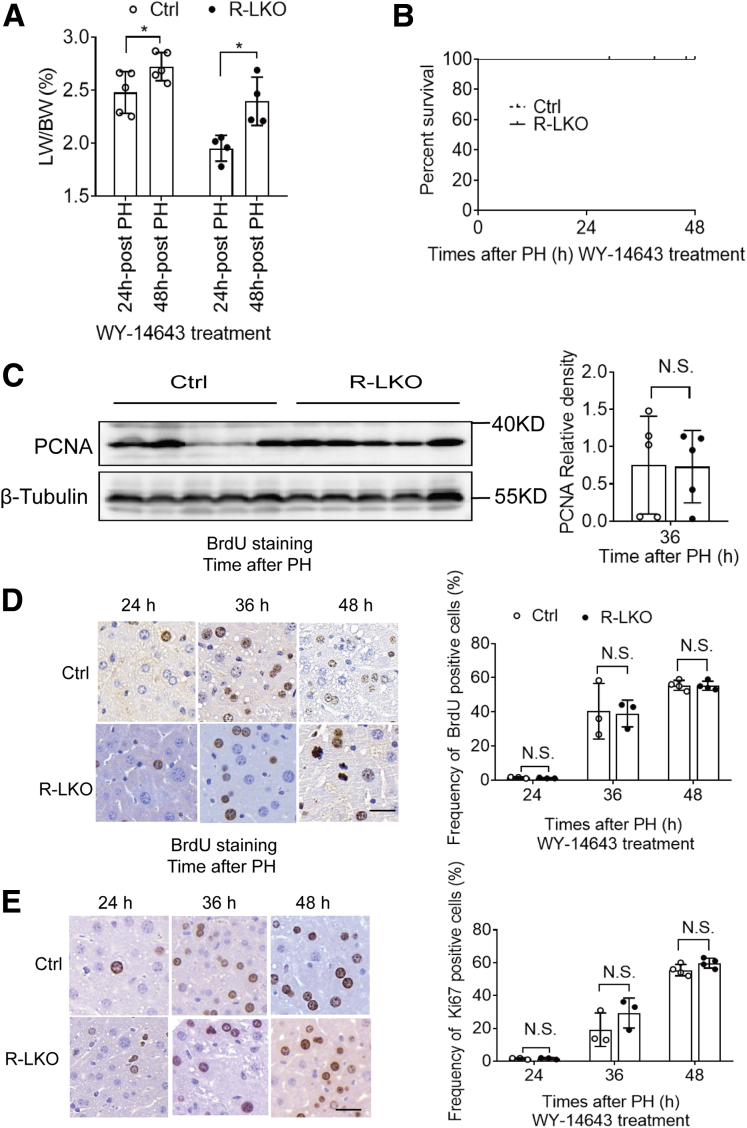
After WY-14643 treatment, the LW/BW in R-LKO mice was increased significantly (P < .05) (Figure 9A), and the survival rates also were increased in R-LKO mice 48 hours after PH (Figure 9B). The expression of PCNA (Figure 9C), as well as BrdU- and Ki-67–positive staining (Figure 9D and E), showed the same levels between R-LKO and the control group after WY-14643 treatment. These results indicated that WY-14643 could enhance liver regeneration and ameliorate survival rates in R-LKO mice. Therefore, the decreased liver regeneration observed in R-LKO mice was caused by inactivation of the PPAR-α pathway.
Figure 9.
WY-14643 improved liver regeneration and survival rate in R-LKO mice. (A) LW/BW of WY-14643– or vehicle-treated mice at the indicated time points after PH. Means ± SD, 4–5 mice per time point. (B) Survival curves from WY-14643–treated control (Ctrl) and R-LKO mice after PH. The survival curves representing the Ctrl (n = 8) and the R-LKO (n = 10) mice were compared using the log-rank test. (C) Western blot analysis of hepatic PCNA expression in WY-14643–treated Ctrl and R-LKO mice at 36 hours after PH. Quantifications were normalized to β-tubulin (n = 5). Means ± SD, 5 mice per time point. (D) Representative immunohistochemistry images for BrdU-positive cells in WY-14643–treated Ctrl and R-LKO mice after PH. Right: Graph shows the quantification of BrdU-positive cells/total cells at the indicated time points after PH. Means ± SD, 3–4 mice per time point. (E) Representative immunohistochemistry images for the Ki-67–positive cells in WY-14643–treated Ctrl and R-LKO mice after PH. Right: Graph shows the quantification of Ki-67–positive cells/total cells at the indicated time points after PH. Means ± SD, 3–5 mice per time point. Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3DHISTECH Ltd). ∗P < .05.
Promotion of the Cell Cycle by Inhibiting the p53 Pathway Could Not Rescue Deficient Liver Regeneration in R-LKO Mice
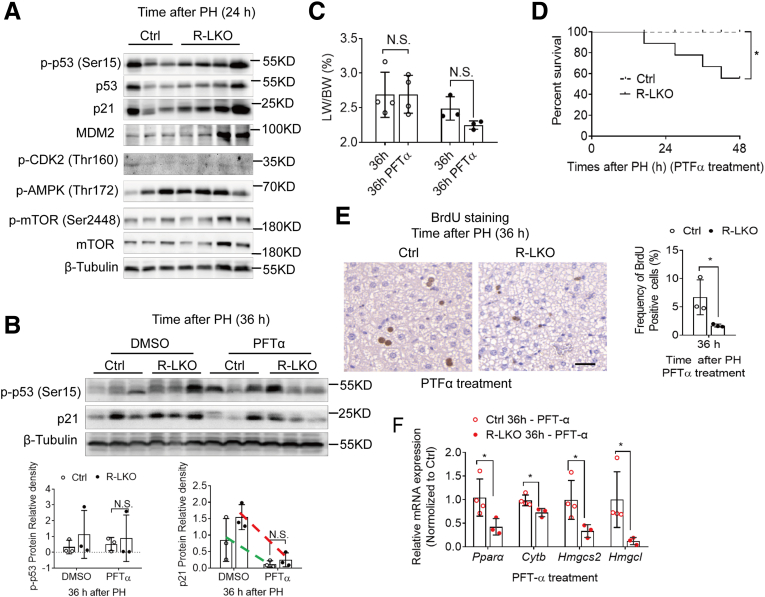
We verified cell-cycle–related proteins in regenerated R-LKO livers. The p53–p21 pathway activity tended to increase in R-LKO–regenerated livers (Figure 10A). To test whether the block in the cell cycle was the cause of insufficient lipid utilization and deficient liver regeneration in R-LKO mice, we promoted cell-cycle progression by treating cells with Pifithrin-α (PFT-α), an inhibitor of p53 target gene transcription. Expression of p21 was inhibited by PFT-α treatment (Figure 10B); the LW/BW and the survival rate in the R-LKO group remained lower than those in the control group after treatment (Figure 10C and D). There was a decrease in the percentage of BrdU-positive cells (Figure 10E) and down-regulated expression of PPAR-α pathway–related genes (Figure 10F) in the R-LKO group, as in pretreatment conditions. These results showed that inhibition of the p53–p21 pathway does not promote hepatocyte proliferation in mTORC2 KO livers without up-regulating FA oxidation.
Figure 10.
The p53 inhibitor could not improve liver proliferation during regeneration. (A) Western blot analysis of hepatic p-p53 (Ser15), p53, p21, mdm2, p-CDK2, Phospho-Adenosine 5’-monophosphate (AMP)-activated protein kinase (p-AMPK [Thr172]), p-mTOR (Ser 2448), mTOR, expression in control (Ctrl) and R-LKO mice 24 hours after PH. (B) Western blot analysis of hepatic p21, p-p53 (Ser15) expression in vehicle and p53 inhibitor Pifithrinα (PTFα)–treated mice 36 hours after PH (3 mice for each time point). Quantifications were normalized to β-tubulin. Green line, control group; red line, R-LKO group. (C) LW/BW of p53 inhibitor PTF-α–treated Ctrl and R-LKO mice 36 hours after PH (3–4 mice for each time point). (D) Survival curves of p53 inhibitor PTF-α–treated Ctrl (n = 9) and R-LKO (n = 9) mice after PH. (E) Representative immunohistochemistry pictures for the BrdU-positive cells in PTF-α–treated Ctrl and R-LKO mice after PH. Right graph: frequency of positive BrdU cells 36 hours after PH (3 mice for each time point). (F) Relative mRNA expression levels of hepatic ketogenesis genes of PTF-α–treated Ctrl and R-LKO mice at the indicated time points. The expression of PPARα, Cytb, Hmgcs2, and Hmgcl are normalized to the levels of control hepatocytes before PH (3–4 mice for each time point). Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3D HISTECH, Ltd). The VersaDoc Imaging System (Bio-Rad) and software were used to visualize proteins. ImageJ was used to quantify the band densities. Data are presented as means ± SD. ∗P < .05. DMSO, dimethyl sulfoxide.
Inactivation of the PPAR-α Pathway in R-LKO Mice Was Dependent on AKT Activation
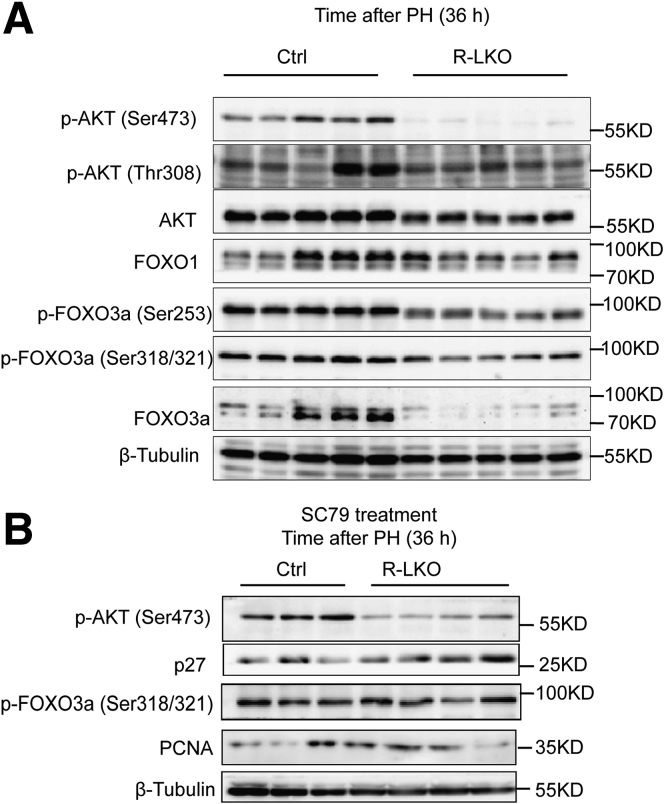
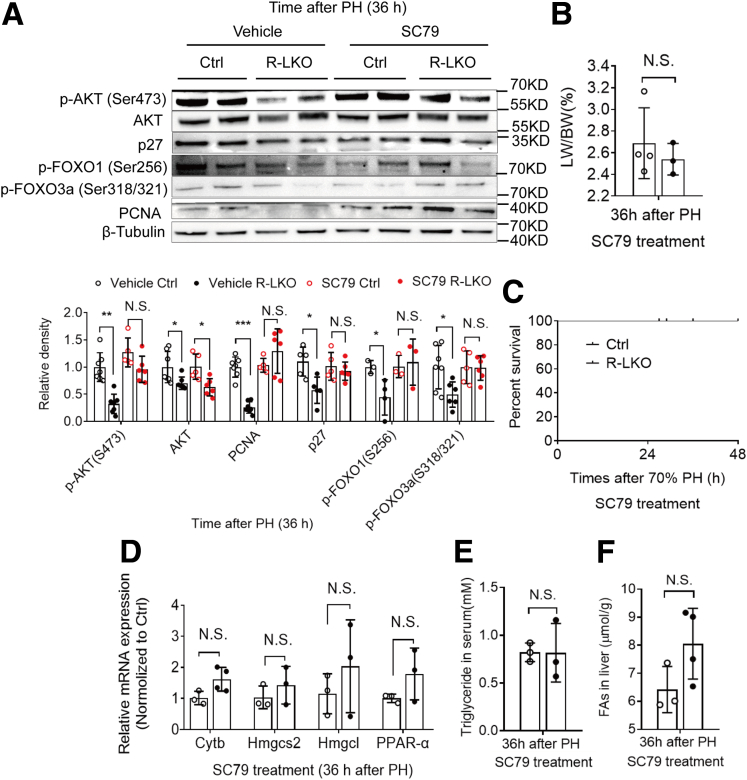
During liver regeneration, AKT pathway activity was decreased significantly (P < .01) in mTORC2-deficient mice (Figure 11A and Figure 12A). To further test the role of AKT in liver regeneration in mTORC2-deficient mice, we treated control and R-LKO mice with the AKT activator SC79. After SC79 treatment, the AKT pathway was significantly (P < .01) activated (Figure 12A and Figure 11A and B). After SC79 treatment, the LW/BW ratio in R-LKO mice showed no difference from that in control group mice 36 hours after PH (Figure 12B). The regenerative ability of R-LKO mice also increased and was comparable with that of the mice in the control group, as indicated by the increased expression of PCNA (Figure 12A). Furthermore, SC79 rescued the survival rate of the R-LKO mice after PH (Figure 12C). SC79 also activated the PPAR-α pathway and decreased TG and FA levels in R-LKO mice (Figure 12D–F). These results suggest that down-regulation of the PPAR-α pathway in R-LKO mice was dependent on AKT pathway activity.
Figure 11.
SC79 could activate AKT pathway in regenerated liver. (A) Western blot analysis of hepatic p-AKT (Ser473), p-AKT (Thr308), AKT, FoxO1, p-FoxO3a (Ser253), p-FoxO3a (Ser318/S321), and FoxO3a expression in control (Ctrl) and R-LKO mice at 36 hours after PH. (B) Western blot analysis of SC79-treated hepatic p-AKT (Ser473), p27, p-FoxO3a (Ser318/S321), and PCNA expression in Ctrl and R-LKO mice at 36 hours after PH. The VersaDoc Imaging System (Bio-Rad) and software were used to visualize proteins.
Figure 12.
AKT agonist restores the PPAR-α pathway and cell proliferation in mTORC2 absent mice liver. (A) Western blot analysis of hepatic phosphorylated protein kinase B [p-AKT (Ser473)], p27, phospho-Forkhead box O3a (p-FoxO3a) (Ser318/S321), FoxO3a, and PCNA expression in vehicle and AKT activator SC79-treated control (Ctrl) and R-LKO mice 36 hours after PH. Quantifications were normalized to β-tubulin. Means ± SD, 3–7 mice per time point. (B) Index of liver to body weight (ILBW) of AKT activator SC79-treated Ctrl and L-RKO mice 36 hours after PH. Means ± SD, 3–4 mice per time point. (C) Survival curves of AKT activator SC79-treated Ctrl (n = 3) and L-RKO (n = 6) mice after PH. (D) Relative mRNA expression levels of hepatic ketogenesis genes of AKT activator SC79-treated Ctrl and L-RKO mice at the indicated time points. The expression of PPARα, Cytb, Hmgcs2, and Hmgcl were normalized to the levels of the control group. Means ± SD, 3–4 mice per group. (E) Analysis of TG contents in serum of AKT activator SC79-treated Ctrl and R-LKO mice 36 hours after PH. Means ± SD, 3 mice per group. (F) Analysis of FA contents in liver of AKT activator SC79-treated Ctrl and R-LKO mice 36 hours after PH. Means ± SD, 3–4 mice per group. The VersaDoc Imaging System (Bio-Rad) and software was used to visualize proteins. ImageJ was used to quantify the band densities.
GluCer Directly Activated PPAR-α and Restored Cell Proliferation in the mTORC2 KO Mouse Liver
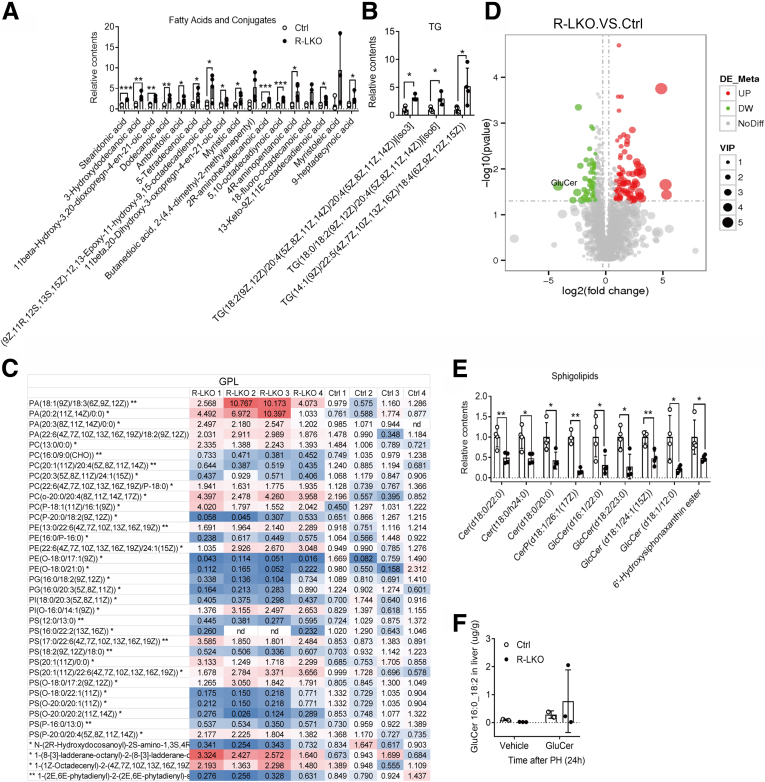
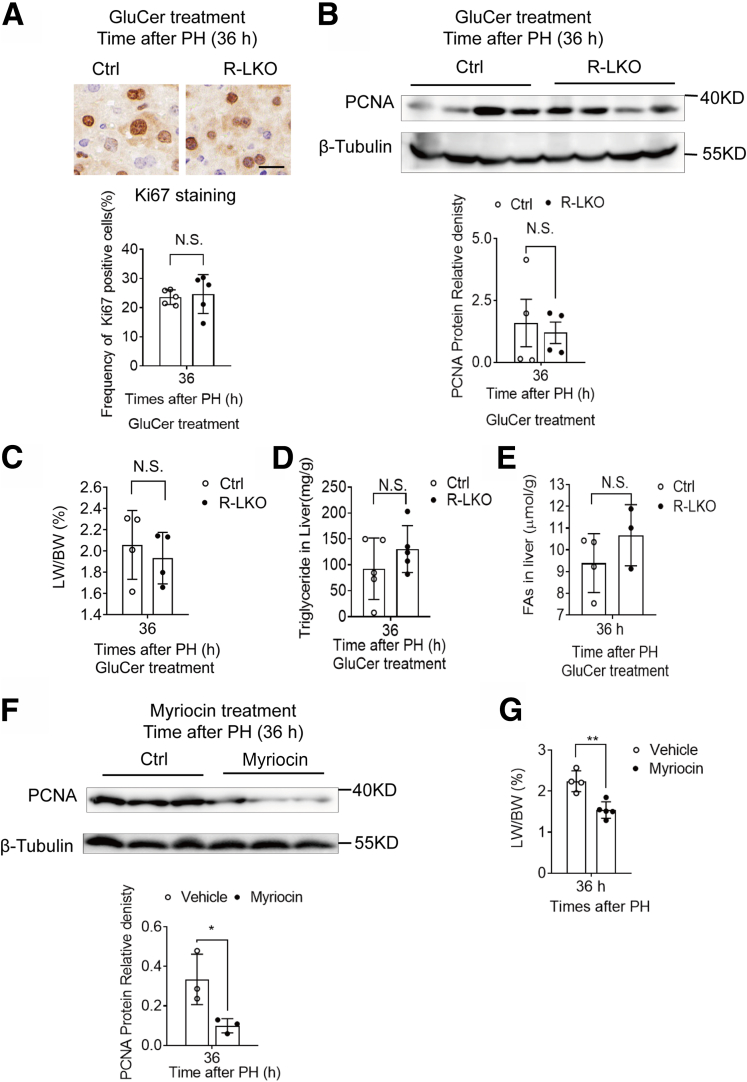
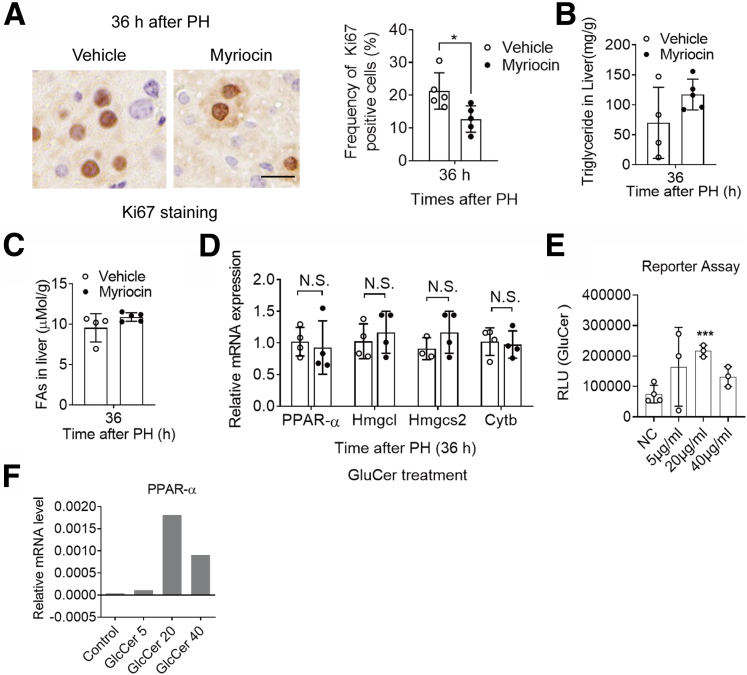
To further investigate the lipid content in regenerated R-LKO livers, we analyzed the lipid content of tissue samples using mass spectrometry at the 24-hour time point. The mostly altered lipid species were FA and conjugates, glycerolipids, in particular, TGs, glycerophospholipids, and sphingolipids (SPs) (Supplementary Table 1). In accordance with our previous results, FA, as well as its conjugates, and TG contents were increased in the mTORC2-deficient liver sample (Figure 13A and B). The altered 37 glycerophospholipids were divided into 2 parts: 18 of 37 lipids such as phosphatidic acid (PA) were increased, and the rest 19 of 37 were decreased (Figure 13C). Specifically, all of the altered SP contents were decreased significantly (P < .05, .01), and the majority were GluCer (Figure 13D and Figure 13E). SPs are synthesized by FAs and ultimately used as signaling molecules or membrane-building blocks. GluCer, a target of mTORC2 in promoting hepatic tumorigenesis,19 was mostly decreased (Figure 13D). To further test the role of SPs in liver regeneration in mTORC2-deficient mice, we treated the control and R-LKO mice with GluCer. Exogenous GluCer treatment increased hepatic GluCer levels in most of the examined animals (4 of 5) (Figure 13F) and eliminated the differences in the LW/BW ratio, Ki-67 positivity, and PCNA, TG, and FA levels between the control and R-LKO groups (Figure 14A and B and Figure 14C–E). Myriocin, an inhibitor of serine palmitoyltransferase (SPT), inhibits de novo SP synthesis. Mice were treated with myriocin 7 days before PH, and hepatocyte proliferation as indicated by PCNA (P < .05), LW/BW (P < .01), and Ki-67 (P < .05) was inhibited significantly 36 hours after PH (Figure 14F and Figure 14G and Figure 15A). The TG and FA contents in myriocin-treated livers showed a tendency to increase, without significant differences (Figure 15B and C). These data indicate that GluCer is essential for hepatocyte proliferation during liver regeneration. After GluCer treatment, the mRNA expression levels of PPAR-α pathway genes, Pparα, Hmgcl, Hmgcs2, and Cytb, were increased in the R-LKO mice at 36 hours after PH (Figure 15D). The fluorescence intensity of the PPAR-α luciferase reporter also was increased after GluCer treatment (Figure 15E), with an increase in PPAR-α mRNA expression after treatment (Figure 15F).
Figure 13.
Decreased SPs in regenerated liver of R-LKO mice. (A) The lipid mass spectrum analysis results of FA and conjugates in regenerated liver of control (Ctrl) and R-LKO mice 24 hours after PH (3–4 mice for each time point). (B) The lipid mass spectrum analysis results of TG contents in regenerated liver of Ctrl and R-LKO mice 24 hours after PH (3–4 mice for each time point). (C) The lipid mass spectrum analysis results of sphingolipids in regenerated liver of Ctrl and R-LKO mice 24 hours after PH (3–4 mice for each time point). (D) The lipid spectrum figure of regenerated liver in control (Ctrl) and R-LKO mice 24 hours after PH. GluCer is the most decreased in content (R-LKO vs Ctrl). (E) The lipid mass spectrum analysis results of glycerophospholipids (GPL) contents in regenerated liver of Ctrl and R-LKO mice 24 hours after PH (3–4 mice for each time point). (F) The lipid mass spectrum analysis results of GluCer (16:0_18:2) contents in regenerated liver of Ctrl and R-LKO mice 24 hours after PH (2–3 mice for each time point). ∗P < .05, ∗∗P < .01 and ∗∗∗P < .001.
Figure 14.
Supplying GluCer restores cell proliferation in mTORC2 KO mice liver. (A) Representative immunohistochemistry images for the Ki-67–positive cells in GluCer-treated Ctrl and R-LKO mice after PH. Right: Graph shows the quantification of Ki-67–positive cells/total cells at the indicated time points after PH. Means ± SD, 5 mice per group. (B) Western blot analysis of hepatic PCNA expression in GluCer-treated Ctrl and R-LKO mice at 36 hours after PH. Quantifications were normalized to β-tubulin. Means ± SD, 4 mice per group. (C) LW/BW of GluCer-treated control (Ctrl) and R-LKO mice 36 hours after PH (3–4 mice for each time point). (D) Triglyceride levels in GluCer-treated Ctrl and R-LKO mice liver 36 hours after PH (3–4 mice for each time point). (E) FA levels in GluCer-treated Ctrl and R-LKO mice liver 36 hours after PH (3 mice for each time point). (F) Western blot analysis of hepatic PCNA expression in myriocin-treated Ctrl and R-LKO mice at 36 hours after PH. Quantifications were normalized to β-tubulin. Means ± SD, 3 mice per group. (G) LW/BW of vehicle and myriocin-treated mice 36 hours after PH (4–5 mice for each time point).
Figure 15.
Supplying GluCer activated PPARα pathway in mTORC2 KO mice liver. (A) Ki-67 staining in vehicle and myriocin-treated mice liver 36 hours after PH (5 mice for each time point). (B) TG levels in vehicle and myriocin mice liver 36 hours after PH (4–5 mice for each time point). (C) FA levels in vehicle and myriocin mice liver 36 hours after PH. (D) Relative mRNA expression levels of hepatic ketogenesis genes of GluCer-treated Ctrl and R-LKO mice at the indicated time points. The expression of PPAR-α, Cytb, Hmgcs2, and Hmgcl were normalized to the levels of control hepatocytes before PH. Means ± SD, 3–4 mice per group. (E) Luciferase reporter assays were performed in GluCer-treated H293T cells, transfected with PPRE-luc and pSG5 PPAR-α. Means ± SD, 3 repeats each group. Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3DHISTECH Ltd). The VersaDoc Imaging System (Bio-Rad) and software was used to visualize proteins. ImageJ was used to quantify the band densities. (F) Relative PPARα mRNA expression in different concentrations in GluCer-treated H293T cells. H293T cells were transfected with the plasmids of peroxisome proliferator activated receptor-response element-luciferase reporter (PPRE-luc) and pSG5 PPARα. Immunohistochemistry slices were scanned and captured by Pannoramic MIDI (3D HISTECH, Ltd). Data are presented as means ± SEM.
Discussion
The role and source of accumulated lipid droplets during liver regeneration are controversial.7,13,23 mTORC2 plays an important role in de novo lipid synthesis. The TG content in the livers of R-LKO mice was decreased significantly under normal conditions.18 However, we found that during liver regeneration, TG and FA contents in the R-LKO mouse livers were increased significantly after PH. Xu et al20 also showed an accumulation of lipid droplets after 3 days. Furthermore, the delayed lipid accumulation was estimated by Oil Red O staining, similar to our study, in which we have used the same technique to evaluate the accumulation tendency and reconfirmed it by 2 other methods, namely, biochemical analyses and lipidomics using liquid chromatography. Liver-specific KO of AKT1/2 results in an antisteatotic effect leading to increased lethality,24 resembling the downstream effects of mTORC2 KO. However, AKT1/AKT2 KO24 might differ from mTORC2 KO in terms of AKT activity. For example, in AKT1/AKT2 KO mice, the activity of both AKT phosphorylated site at serine 473 (pSer473) and AKT phosphorylated site at threonine 308 (pThr308) is lost, whereas in mTORC2 KO mice, only AKT pSer473 activity is almost lost. In line with this notion, only AKT (Ser473) was reported to rescue fatty acid synthesis in R-LKO mice. AKT Thr308 activated by pyruvate dehydrogenase kinase 1 (PDK1). PDK1 could phosphorylate ribosomal protein S6 kinase (S6K) and mTORC1,25 inhibit which could strongly provoke PPAR-α ketogenesis pathway,26,27 which might be a main cause of antisteatotic effect.28
Our data also showed that mRNA expression levels of de novo FA synthesis genes, such as Fasn, Acly, and Srebp1c, were decreased significantly in regenerated R-LKO livers 36 hours after PH, which is the same time point at which lipid accumulation also occurred. We concluded that de novo FA synthesis did not correlate with TG accumulation. These results are consistent with normal liver regeneration in Fasn KO mice.13 Furthermore, the expression of the lipid transport-related genes Cd36 and Fabp4 peaked at 24 hours after PH. To ensure the effective use of lipid droplets, the PPAR-α–FA oxidation pathway also peaked at 24 hours. However, lipid overloading delays DNA replication in hepatocytes.9 To overcome this side effect, lipid export-related genes such as Mttp21 also showed a peak in expression after lipid oxidation to remove extra fat after supplying energy. This removal might be downstream of the PPAR-α pathway and not the signal derived from lipid overload. As in R-LKO mice, despite lipid overloading, the Mttp genes remained silent, similar to PPAR-α pathway genes. Moreover, in the control group, the genes responsible for hepatocyte transport were up-regulated after activation of the PPAR-α pathway. Consequently, it can be concluded that lipid metabolism is a fine-tuned process during liver regeneration. Overall, TGs and FAs are transported into hepatocytes, and the PPAR-α pathway metabolizes FAs to provide energy for hepatocyte proliferation. These results are consistent with previously obtained findings that cluster of differentiation 36 contributes to the enhancement of transient regeneration-associated steatosis.16 Hepatocytes are selected to increase lipid transport rather than synthesis, which requires more energy and raw materials, as a survival mode in emergencies such as PH.
Liver regeneration is a highly precise and efficient process.29 Hepatocyte size starts increasing after PH, reaches a peak value at 36 hours, and then returns to normal 96 hours after PH.30 The hepatocyte population also increases from 36 hours onward after PH and is sustained until the end of liver regeneration.30 Reconstruction requires high bioenergy to ensure liver cell repopulation. However, the mechanism of metabolic pattern transformation during different stages of liver regeneration remains unclear. Glucose levels decreased significantly during liver regeneration after PH, similar to the results of previous studies.30 Oxidative phosphorylation levels have been reported to decrease after PH31; accordingly, during liver regeneration, increased cell size is accompanied by down-regulation of mitochondrial and lipid biosynthesis-related genes. Furthermore, the glycolysis rate increases with cell size.30,32,33 For lipid metabolism, lipid droplets started to accumulate 12 hours after PH, reaching a peak value at 36 hours, and were reduced to the basal level during the advanced stage of liver regeneration. Systemic knockdown of PPAR-α causes a 12- to 24-hour lag at the G1/S checkpoint,14 whereas liver-specific deletion of PPAR-α significantly reduces hepatocyte proliferation at 32 hours.15 Acceleration of lipid metabolism by a PPAR-α agonist before PH accelerates liver regeneration.34 In accordance with previous findings, the PPAR-α activator WY14643 significantly increased liver size and proliferation during liver regeneration.35 In summary, we hypothesized that glycolysis is a pivotal energy source for the early cell size growth phase, and FA oxidation is a major energy source for the increase in both hepatocyte size and proliferation.
Fatty acid oxidation is an essential factor that promotes cell proliferation during liver regeneration. PFT-α treatment every 48 hours after PH only inhibited p53 expression in the nucleus and had no effect on p21 expression.36 Conversely, daily pretreatment of mice with the p53 inhibitor PFT-α 1 week before PH significantly reduced the levels of p21 protein in hepatocytes.37 However, acceleration of the cell cycle by p21 inhibition did not promote FA oxidation and could not drive hepatocyte proliferation and liver regeneration. Consistently, it did not rescue the survival rate of the R-LKO mice. In turn, the PPAR-α agonist increased the oxidation of accumulated lipids in the mTORC2-deficient liver during liver regeneration. Finally, improvement in lipid oxidation promoted hepatocyte proliferation and rescued survival rate.
Previous studies have shown that FAs and their metabolites are activators of the PPAR-α signaling pathway. Considering the accumulated FA and TG contents and depressed PPAR-α activation in R-LKO livers after PH, a lipidomics analysis was performed to screen for potential activators of PPAR-α. The most down-regulated SPs were considered. SPT is a key enzyme in SP synthesis, and liver-specific SPT KO mice show disrupted liver regeneration after PH.38 However, the role of GluCer (one of the terminal products of SP synthesis) during liver regeneration remains unclear because the GluCer content showed no change in SPT-deficient livers. GluCer is a key factor for mTORC2 that induces tumorigenesis.19 Furthermore, the SPT inhibitor myriocin significantly decreased GluCer levels in hepatic tumors19 and inhibited hepatocyte cell proliferation. In addition, our study showed that GluCer could activate the PPAR-α pathway and significantly rescue the controlled proliferation in the mTORC2-deficient livers after PH.
mTORC2 regulates glucose and lipid metabolism through AKT.18 In turn, AKT has been reported to facilitate hepatocyte proliferation by inhibiting Forkhead box O1 expression24; however, its relationship with PPAR-α currently is unknown. We showed that after SC79 (an AKT activator) treatment in R-LKO mice, the PPAR-α pathway also was activated, possibly because of the promotion of GluCer synthesis by SC79, which is supported by evidence from human liver biopsy specimens showing that AKT (Ser473) activity is correlated positively with GluCer level.19 Therefore, we speculated that the down-regulation of the PPAR-α pathway in the R-LKO liver is dependent on AKT activation.
Therefore, using R-LKO mice in the present study, we elucidated the mechanisms involved in lipid metabolism and function during liver regeneration. Furthermore, we showed that the generalized lipid accumulation observed in the mTORC2-deficient liver during regeneration after PH was associated with down-regulation of the PPAR-α pathway. Conversely, we showed that enhanced lipid metabolism by GluCer through the activation of the PPAR-α pathway can improve liver regeneration, and that GluCer may be a key factor associated with the activation of mTORC2 and PPAR-α during liver regeneration.
Materials and Methods
Animals
Generation of Rictor conditional KO (Rictorflox/flox) mice has been described previously.18 Rictorflox/flox mice were crossed with albumin-Cre (provided by Lu Zhongjie) mice to generate LKO mice with the genotype Rictorflox/flox–albumin-Cre mice (R-LKO). Rictorflox/flox mice were used as the littermate control. All animals received humane care according to the Animal Research: Reporting of In Vivo Experiments guidelines, and all animal experiments were approved by the Animal Care and Ethics Committee of Jinan University. Animals used for the experiments were 8- to 12-week-old males or females. All mice had a C57 background. Both male and female mice were used indistinctively. Two thirds of the PH procedures were performed according to a previously described method, the left and median lobes were resected for detail.39,40 The PPAR-α activator WY-14643 (S8029; Selleck) was administered at 50 mg/kg/d 2 weeks before PH. For GluCer (131304P; Sigma-Aldrich) administration, animals received daily intraperitoneal injections of 1.5 μg/dose for 3 days before PH.41 Animals were injected with the p53 inhibitor, pifithrin-α (S2929, 2.2 mg/kg; Selleck), or control vehicle, dimethyl sulfoxide, dissolved in physiological saline 3 times per week for 2 weeks before PH. SC79 (S7863; Selleck) was applied via intraperitoneal injection at a concentration of 0.04 mg/g of body weight, and dimethyl sulfoxide was used as a control vehicle. Myriocin (HY-N6798; Med Chem Express) was administered intraperitoneally to 8-week-old R-LKO and control mice at a dosage of 0.3 mg/kg, every alternate day for 2 weeks. All animals were injected intraperitoneally with 100 mg/kg body weight BrdU (B9285; Sigma-Aldrich) every day, along with 0.8 mg/mL BrdU water before PH until 2 hours before they were killed. Liver remnants were removed, weighed, snap-frozen in liquid nitrogen, or immediately processed for histologic analysis.
Real-Time Quantitative Polymerase Chain Reaction Analysis
Total RNA was isolated from liver samples using TRIzol (15596-026; Invitrogen), reverse-transcribed, and analyzed using real-time quantitative polymerase chain reaction with the SYBR Green master mix (1725272; Bio-Rad) and target-specific primers (Table 1) on a Bio-Rad CFX96 Touch. Liver complementary DNA samples were pooled from 3 mice per group at 24 and 36 hours after PH.
Table 1.
Primer Table
| Gene | Primer sequence, position on mRNA |
|---|---|
| SREBPIc | F: 5'-GGAGCCATGGATTGCACATT-3' |
| R: 5'-GGCCCGGGAAGTCACTGT-3' | |
| Fasn | F: 5'-TGGTGGTGTGGACATGGTCACAGA-3' |
| R: 5'-CCGAAGCTGGGGGTCCATTGTGTG-3' | |
| Elovl5 | F: 5'-GAACATTTCGATGCGTCACTCA-3' |
| R: 5'-GGAGGAACCATCCTTTGACTCTT-3' | |
| FABP4 | F: 5'-AAGGTGAAGAGCATCATAACCCT-3' |
| R: 5'-TCACGCCTTTCATAACACATTCC-3' | |
| MTTP | F: 5'-GAGCGGCTATACAAGCTCACGTAC-3' |
| R: 5'-TCACCATCAGGATTCCTCCACAGT-3' | |
| Actin | F: 5'-GAC ATGGAGAAGATCTGGCA-3' |
| R: 5'-GGTCTCAAACATGATCTGGGT-3' | |
| Acaca | F: 5'-GGATGACAGGCTTGCAGCTAT-3' |
| R: 5'-TTTGTGCAACTAGGAACGTAAGTCG-3' | |
| Acox1 | F: 5'-CTGCCAAGGGACTCCAGAGCAGCT-3' |
| R: 5'-GACATGGACACATCCACCATGCAG-3' | |
| PPARɑ | F: 5'-TCTCCACGTTCCAGCCCTTCCTCA-3' |
| R: 5'-TTCACATGCGTGAACTCCGTAGTG-3' | |
| CytB | F: 5'-ATTCCTTCATGTCGGACGAG-3' |
| R: 5'-ACTGAGAAGCCCCCTCAAAT-3' | |
| Hmgcs2 | F: 5'-AGAGAGCGATGCAGGAAACTT-3' |
| R: 5'-AAGGATGCCCACATCI I I IGG-3' | |
| Hmgcl | F: 5'-CCGGCATCAACTACCCAGTC-3' |
| R: 5'-GCGCTGGAAACTCTCCTCTAT-3' | |
| Dgat2 | F: 5'-AGTGGCAATGCTATCATCATCGT-3' |
| R: 5'-AAGGAATAAGTGGGAACCAGA-3' | |
| CD36 | F: 5'-GCCAAGCTATTGCGACATGAT-3' |
| R: 5'-CAGATCCGAACACAGCGTAGA-3' | |
| Slc27a2 | F: 5'-ATCGTGGTTGGGGCTACTTTAG-3' |
| R: 5'-TTTGGTTTCTGCGGTGTGTTG-3' | |
| Slc27a5 | F: 5'-GAGGGCAATGTGGGCTTAATG-3' |
| R: 5'-AGGCTCTGCTGTCTCTATGTC-3' | |
| Acc1 | F: 5'-AAGGCTATGTGAAGGATG-3' |
| R: 5'-CTGTCTGAAGAGGTTAGG-3' | |
| Acc2 | F: 5'-CTTGCTTCTCTTTCTGACTTG-3' |
| R: 5'-GGCTTCCACCTTACTGTTG-3' | |
| Acly | F: 5'-TGTTCTTGGTCAGCTTTGTAGC-3' |
| R: 5'-AGGCTGTGGGTCTTGTTTAGG-3' |
F, forward; mRNA, messenger RNA; R, reverse.
Liver Histology and Immunohistochemical Analyses
Liver tissues were fixed in 0.01 mol/L phosphate-buffered saline (pH 7.4) containing 10% formalin, embedded in paraffin, sectioned, and stained with H&E for histologic examination. Immunohistochemistry was performed with mouse anti-BrdU (B2531; Sigma-Aldrich) and Ki-67 (9129S; CST) antibodies at 1:1500 dilution. For Oil Red O staining, the freshly isolated tissues were fixed in optimal cutting temperature compound and sliced at −20°C. Frozen sections were fixed in cold methanol, rinsed with 60% isopropanol, and stained for 20 minutes with Oil Red O solution (E607319; Sangon Biotech) at 37°C. After 2 rinses with distilled water, the slides were stained with hematoxylin for 1 minute, rinsed with distilled water, and mounted. Ten fields per section from 3 sections per mouse were randomly chosen and examined.
Western Blot Analysis
Liver tissues were homogenized and lysed in radio immunoprecipitation assay buffer (P0013B; Beyotime, Co) containing protease and phosphatase inhibitors. The liver tissue homogenate was centrifuged at 15,000 × g at 4°C, and the supernatant was collected. Protein concentration was measured using the bicinchoninic acid method. Proteins were separated on 8% and 12% sodium dodecyl sulfate–polyacrylamide gels and transferred to 0.2-μm nitrocellulose membranes for 100 minutes at 4°C. The antibodies used are listed in Table 2 and were incubated overnight at 4°C. This was followed by incubation with the secondary goat anti-rabbit- (Ultra Sensitive Chemiluminescent Immunoassay Substrate) horseradish-peroxidase antibody at a 1:3000 dilution in 5% bovine serum albumin in Tris buffered saline Tween (TBST) and developed with enhanced chemiluminescence Western chemiluminescent horseradish-peroxidase substrate. The Bio-Rad VersaDoc imaging system and software were used to visualize the proteins. Signal intensities were quantified and normalized to tubulin levels using the ImageJ Pro Plus software (National Institutes of Health, Bethesda, MD).
Table 2.
Antibodies Used in Western Blot
| Antibody | Dilution | Source | Catalog number |
|---|---|---|---|
| PCNA | 1:2000 | Cell Signaling Technology | 13110 |
| FASN | 1:1000 | Cell Signaling Technology | 3180 |
| DGAT1 | 1:500 | BOSTER Biological Technology | PB0419 |
| ACLY | 1:1000 | Cell Signaling Technology | 13390 |
| CD36 | 1:500 | Proteintech | 18836-1-AP |
| MTTP | 1:1000 | Santa Cruz | sc-135994 |
| p21 | 1:500 | Santa Cruz | sc-6246 |
| p-p53 (Ser 15) | 1:1000 | Cell Signaling Technology | 9284 |
| Mdm2 | 1:1000 | Cell Signaling Technology | 51541 |
| P-CDK2 | 1:1000 | Cell Signaling Technology | 2561 |
| p-AMPK (Thrl72) | 1:1000 | Cell Signaling Technology | 8208 |
| p-AKT (Ser473) | 1:1000 | Cell Signaling Technology | 4046 |
| p-AKT (Thr308) | 1:1000 | Cell Signaling Technology | 13038 |
| p-mTOR (Ser2448) | 1:1000 | Cell Signaling Technology | 5536 |
| mTOR | 1:1000 | Cell Signaling Technology | 2983 |
| β-tubulin | 1:5000 | Cell Signaling Technology | 2148 |
| FoxO1 | 1:1000 | Cell Signaling Technology | 2880 |
| P-Fox03a (S253) | 1:1000 | Cell Signaling Technology | 9466 |
| P-Fox03a (S318/S321) | 1:1000 | Cell Signaling Technology | 9465 |
| p27 | 1:500 | Santa Cruz | sc-1641 |
Measurement of Triglycerides and Free Fatty Acid in the Serum and Liver Tissue
For the serum samples, reagents for biochemical analyses were purchased from DiaSys Diagnostic Systems GmbH (Holzheim, Germany). Serum samples were tested on a HITACHI Clinical Analyzer 7180 according to the manufacturer’s protocol. For liver tissue, the extraction and tests were performed using Biovision-free fatty acid (K612-100) and triglyceride (K622-100) quantification kits. Liver tissues (100 mg) were homogenized in 1 mL solution containing 5% Nonidet P-40 in water, slowly heated in a water bath at 80°C–100°C until the liquid became cloudy, followed by cooling. This process was repeated 2 or 3 times. The samples then were centrifuged at 15,000 × g for 2 minutes, and the supernatant was collected.
Liquid Chromatography Analysis for Lipidomics
Samples were injected onto a C18 Charged Surface Hybrid column (100 mm × 2.1 mm, 1.7 μm; Waters, Manchester, UK) using a 20-minute linear gradient at a flow rate of 0.4 mL/min. The column temperature was set to 60°C. The mobile phase buffer A was constitute by 60% water and 40% acetonitrile, whereas buffer B was constitute by 10% acetonitrile isopropanol (1/9), with 10 mmol/L ammonium formate and 0.1% formic acid in each solution. The solvent gradient was set as follows: 40% B, initial; 43% B, 2 minutes; 50% B, 2.1 minutes; 54% B, 12 minutes; 70% B, 12.1 minutes; 99% B, 18 minutes; and re-equilibration for 2 minutes at 40% B. After separation using ultra performance liquid chromatography, mass spectrometry (MS) was performed using a Xevo G2-S Q-TOF instrument with an electrospray ionization source (Waters). Dynamic range enhancement was implemented in the MS method using Xevo G2-S Q-TOF to improve isotopic distribution and mass accuracy and reduce high ion intensities. In the positive-ion mode, MS parameters were as follows: capillary voltage, 2.5 kV; cone voltage, 24 V; source temperature, 100°C; desolvation temperature, 400°C; desolvation gas flow, 800 L/h; and cone gas flow, 50 L/h. Acquisition was performed from 100 to 1500 daltons. In the negative-ion mode, MS parameters were as follows: capillary voltage, 2.5 kV; cone voltage, 25 V; source temperature, 100°C; desolvation temperature, 500°C; desolvation gas flow, 600 L/h; and cone gas flow, 10 L/h. Acquisition was performed from 100 to 1500 daltons (Novogene Co., Ltd). The raw data were imported to Progenesis QI (Waters) for peak alignment to obtain a peak list containing the retention time, mass-to-charge ratio (m/z), and the peak area of each sample.
Transactivation Assays
We co-transfected the PPRE X3-TK-luc (1015; Addgene) and pSG5 PPAR-α (22751; Addgene) plasmid using Lipo8000 (C0533; Beyotime) into HEK293T cells according to the Lipo8000 protocol; then, we added appropriate concentrations of GluCer (131304P; Sigma-Aldrich). We used a dual-luciferase reporter assay system (E1910; Promega) and a MIX Microtiter plate luminometer (Thermo Scientific) to determine luciferase activity in cell lysates.
Statistical Analyses
Statistical significance analyses were performed using the Student t test, the Wilcoxon test, and the log-rang test. Differences with calculated P values less than .05 were considered statistically significant. For Western blot, we used the Image Pro-Plus software to analyze the density of the bands, which were obtained from the Western blot (WB) experiments.
All authors had access to the study data and have reviewed and approved the final manuscript.
Acknowledgments
CRediT Authorship Contributions
Lingling Zhang (Conceptualization: Lead; Investigation: Supporting; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Yanqiu Li (Investigation: Lead)
Ying Wang (Investigation: Lead)
Yugang Qiu (Methodology: Equal; Writing – original draft: Equal)
Hanchuan Mou (Investigation: Equal)
Yuanyao Deng (Investigation: Equal)
Jiyuan Yao (Investigation: Equal)
Zhiqing Xia (Investigation: Equal)
Wenzhe Zhang (Investigation: Equal)
Di Zhu (Investigation: Equal)
Zeyu Qiu (Data curation: Equal)
Zhongjie Lu (Methodology: Equal)
Jirong Wang (Methodology: Equal)
Zhouxin Yang (Methodology: Equal)
GenXiang Mao (Methodology: Supporting)
Dan Chen (Validation: Equal)
Leimin Sun (Project administration: Supporting)
Leiming Liu (Conceptualization: Equal; Investigation: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Zhenyu Ju (Conceptualization: Lead; Funding acquisition: Lead)
Data Availability Statement
All data, analytic methods, and study materials have been made available in the results, figures, and materials and methods of this article.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by National Natural Science Foundation of China grants 92049304, 81801372, 91949125, 82171545, and 82030039; National Key R&D Program of China grant 2021YFA0804903; Program for Guangdong Introducing Innovative and Entrepreneurial Teams grant 2017ZT07S347; and Innovative Team Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory grant 2018GZR110103002. Writing Assistance was provided by the National Natural Science Foundation of China grant 91949125.
Contributor Information
Lingling Zhang, Email: linglingzhang@zju.edu.cn.
Leiming Liu, Email: liuleiming@zju.edu.cn.
Zhenyu Ju, Email: zhenyuju@163.com.
Supplementary Material
References
- 1.Michalopoulos G.K., Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18:40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 2.Jung Y.S., Stratton S.A., Lee S.H., Kim M.J., Jun S., Zhang J., Zheng B., Cervantes C.L., Cha J.H., Barton M.C., Park J.I. TMEM9-v-ATPase activates Wnt/beta-catenin signaling via APC lysosomal degradation for liver regeneration and tumorigenesis. Hepatology. 2021;73:776–794. doi: 10.1002/hep.31305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campana L., Esser H., Huch M., Forbes S. Liver regeneration and inflammation: from fundamental science to clinical applications. Nat Rev Mol Cell Biol. 2021;22:608–624. doi: 10.1038/s41580-021-00373-7. [DOI] [PubMed] [Google Scholar]
- 4.He L., Pu W., Liu X., Zhang Z., Han M., Li Y., Huang X., Han X., Li Y., Liu K., Shi M., Lai L., Sun R., Wang Q.D., Ji Y., Tchorz J.S., Zhou B. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science. 2021;371 doi: 10.1126/science.abc4346. [DOI] [PubMed] [Google Scholar]
- 5.Riddiough G.E., Jalal Q., Perini M.V., Majeed A.W. Liver regeneration and liver metastasis. Semin Cancer Biol. 2021;71:86–97. doi: 10.1016/j.semcancer.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa M., Brooks A.J., Fernandez-Rojo M.A., Medina J., Chhabra Y., Minami S., Tunny K.A., Parton R.G., Vivian J.P., Rossjohn J., Chikani V., Ramm G.A., Ho K.K.Y., Waters M.J. Growth hormone stops excessive inflammation after partial hepatectomy, Allowing liver regeneration and survival through induction of H2-Bl/HLA-G. Hepatology. 2021;73:759–775. doi: 10.1002/hep.31297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez M.A., Albor C., Ingelmo-Torres M., Nixon S.J., Ferguson C., Kurzchalia T., Tebar F., Enrich C., Parton R.G., Pol A. Caveolin-1 is essential for liver regeneration. Science. 2006;313:1628–1632. doi: 10.1126/science.1130773. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Rojo M.A., Restall C., Ferguson C., Martel N., Martin S., Bosch M., Kassan A., Leong G.M., Martin S.D., McGee S.L., Muscat G.E., Anderson R.L., Enrich C., Pol A., Parton R.G. Caveolin-1 orchestrates the balance between glucose and lipid-dependent energy metabolism: implications for liver regeneration. Hepatology. 2012;55:1574–1584. doi: 10.1002/hep.24810. [DOI] [PubMed] [Google Scholar]
- 9.DeAngelis R.A., Markiewski M.M., Taub R., Lambris J.D. A high-fat diet impairs liver regeneration in C57BL/6 mice through overexpression of the NF-kappaB inhibitor, IkappaBalpha. Hepatology. 2005;42:1148–1157. doi: 10.1002/hep.20879. [DOI] [PubMed] [Google Scholar]
- 10.Rudnick D.A. Trimming the fat from liver regeneration. Hepatology. 2005;42:1001–1003. doi: 10.1002/hep.20931. [DOI] [PubMed] [Google Scholar]
- 11.Kohjima M., Tsai T.H., Tackett B.C., Thevananther S., Li L., Chang B.H., Chan L. Delayed liver regeneration after partial hepatectomy in adipose differentiation related protein-null mice. J Hepatol. 2013;59:1246–1254. doi: 10.1016/j.jhep.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamano M., Ezaki H., Kiso S., Furuta K., Egawa M., Kizu T., Chatani N., Kamada Y., Yoshida Y., Takehara T. Lipid overloading during liver regeneration causes delayed hepatocyte DNA replication by increasing ER stress in mice with simple hepatic steatosis. J Gastroenterol. 2014;49:305–316. doi: 10.1007/s00535-013-0780-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newberry E.P., Kennedy S.M., Xie Y., Luo J., Stanley S.E., Semenkovich C.F., Crooke R.M., Graham M.J., Davidson N.O. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology. 2008;48:1097–1105. doi: 10.1002/hep.22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson S.P., Yoon L., Richard E.B., Dunn C.S., Cattley R.C., Corton J.C. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology. 2002;36:544–554. doi: 10.1053/jhep.2002.35276. [DOI] [PubMed] [Google Scholar]
- 15.Xie G., Yin S., Zhang Z., Qi D., Wang X., Kim D., Yagai T., Brocker C.N., Wang Y., Gonzalez F.J., Wang H., Qu A. Hepatocyte peroxisome proliferator-activated receptor alpha enhances liver regeneration after partial hepatectomy in mice. Am J Pathol. 2019;189:272–282. doi: 10.1016/j.ajpath.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y., Tran M., Wang L., Shin D.J., Wu J. PDK4-deficiency reprograms intrahepatic glucose and lipid metabolism to facilitate liver regeneration in mice. Hepatol Commun. 2020;4:504–517. doi: 10.1002/hep4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiwara A., Cornu M., Cybulski N., Polak P., Betz C., Trapani F., Terracciano L., Heim M.H., Ruegg M.A., Hall M.N. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Guri Y., Colombi M., Dazert E., Hindupur S.K., Roszik J., Moes S., Jenoe P., Heim M.H., Riezman I., Riezman H., Hall M.N. mTORC2 promotes tumorigenesis via lipid synthesis. Cancer Cell. 2017;32:807–823 e12. doi: 10.1016/j.ccell.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Xu M., Wang H., Wang J., Burhan D., Shang R., Wang P., Zhou Y., Li R., Liang B., Evert K., Utpatel K., Xu Z., Song X., Che L., Calvisi D.F., Wang B., Chen X., Zeng Y., Chen X. mTORC2 signaling is necessary for timely liver regeneration after partial hepatectomy. Am J Pathol. 2020;190:817–829. doi: 10.1016/j.ajpath.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodson L., Gunn P.J. The regulation of hepatic fatty acid synthesis and partitioning: the effect of nutritional state. Nat Rev Endocrinol. 2019;15:689–700. doi: 10.1038/s41574-019-0256-9. [DOI] [PubMed] [Google Scholar]
- 22.Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 23.Abshagen K., Degenhardt B., Liebig M., Wendt A., Genz B., Schaeper U., Stumvoll M., Hofmann U., Frank M., Vollmar B., Kloting N. Liver-specific Repin1 deficiency impairs transient hepatic steatosis in liver regeneration. Sci Rep. 2018;8 doi: 10.1038/s41598-018-35325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauta M., Rotllan N., Fernandez-Hernando A., Langhi C., Ribera J., Lu M., Boix L., Bruix J., Jimenez W., Suarez Y., Ford D.A., Baldan A., Birnbaum M.J., Morales-Ruiz M., Fernandez-Hernando C. Akt-mediated foxo1 inhibition is required for liver regeneration. Hepatology. 2016;63:1660–1674. doi: 10.1002/hep.28286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez A., Hall M.N., Lin S.C., Hardie D.G. AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell Metab. 2020;31:472–492. doi: 10.1016/j.cmet.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Sengupta S., Peterson T.R., Laplante M., Oh S., Sabatini D.M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 27.Kim K., Pyo S., Um S.H. S6 kinase 2 deficiency enhances ketone body production and increases peroxisome proliferator-activated receptor alpha activity in the liver. Hepatology. 2012;55:1727–1737. doi: 10.1002/hep.25537. [DOI] [PubMed] [Google Scholar]
- 28.Mora A., Lipina C., Tronche F., Sutherland C., Alessi D.R. Deficiency of PDK1 in liver results in glucose intolerance, impairment of insulin-regulated gene expression and liver failure. Biochem J. 2005;385:639–648. doi: 10.1042/BJ20041782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43(Suppl 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 30.Caldez M.J., Van Hul N., Koh H.W.L., Teo X.Q., Fan J.J., Tan P.Y., Dewhurst M.R., Too P.G., Talib S.Z.A., Chiang B.E., Stunkel W., Yu H., Lee P., Fuhrer T., Choi H., Bjorklund M., Kaldis P. Metabolic remodeling during liver regeneration. Dev Cell. 2018;47:425–438 e5. doi: 10.1016/j.devcel.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 31.Vendemiale G., Guerrieri F., Grattagliano I., Didonna D., Muolo L., Altomare E. Mitochondrial oxidative phosphorylation and intracellular glutathione compartmentation during rat liver regeneration. Hepatology. 1995;21:1450–1454. [PubMed] [Google Scholar]
- 32.Clerici E., Ciccarone P. Crabtree effect and the anaerobic glycolysis of the regenerating rat liver. Nature. 1965;207:762–763. doi: 10.1038/207762a0. [DOI] [PubMed] [Google Scholar]
- 33.Forni E., Filipazzi A. [Study of some aspects of liver regeneration after partial hepatectomy. I. Oxygen consumption and glycolysis of regenerating tissue] Chir Patol Sper. 1964;12:263–267. [PubMed] [Google Scholar]
- 34.Aoyama T., Ikejima K., Kon K., Okumura K., Arai K., Watanabe S. Pioglitazone promotes survival and prevents hepatic regeneration failure after partial hepatectomy in obese and diabetic KK-A(y) mice. Hepatology. 2009;49:1636–1644. doi: 10.1002/hep.22828. [DOI] [PubMed] [Google Scholar]
- 35.Fan S., Gao Y., Qu A., Jiang Y., Li H., Xie G., Yao X., Yang X., Zhu S., Yagai T., Tian J., Wang R., Gonzalez F.J., Huang M., Bi H. YAP-TEAD mediates peroxisome proliferator-activated receptor alpha-induced hepatomegaly and liver regeneration in mice. Hepatology. 2022;75:74–88. doi: 10.1002/hep.32105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eipel C., Schuett H., Glawe C., Bordel R., Menger M.D., Vollmar B. Pifithrin-alpha induced p53 inhibition does not affect liver regeneration after partial hepatectomy in mice. J Hepatol. 2005;43:829–835. doi: 10.1016/j.jhep.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Inoue Y., Tomiya T., Yanase M., Arai M., Ikeda H., Tejima K., Ogata I., Kimura S., Omata M., Fujiwara K. p53 May positively regulate hepatocyte proliferation in rats. Hepatology. 2002;36:336–344. doi: 10.1053/jhep.2002.34942. [DOI] [PubMed] [Google Scholar]
- 38.Li Z., Kabir I., Jiang H., Zhou H., Libien J., Zeng J., Stanek A., Ou P., Li K.R., Zhang S., Bui H.H., Kuo M.S., Park T.S., Kim B., Worgall T.S., Huan C., Jiang X.C. Liver serine palmitoyltransferase activity deficiency in early life impairs adherens junctions and promotes tumorigenesis. Hepatology. 2016;64:2089–2102. doi: 10.1002/hep.28845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell C., Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L., Liu L., He Z., Li G., Liu J., Song Z., Jin H., Rudolph K.L., Yang H., Mao Y., Zhang L., Zhang H., Xiao Z., Ju Z. Inhibition of wild-type p53-induced phosphatase 1 promotes liver regeneration in mice by direct activation of mammalian target of rapamycin. Hepatology. 2015;61:2030–2041. doi: 10.1002/hep.27755. [DOI] [PubMed] [Google Scholar]
- 41.Zigmond E., Preston S., Pappo O., Lalazar G., Margalit M., Shalev Z., Zolotarov L., Friedman D., Alper R., Ilan Y. Beta-glucosylceramide: a novel method for enhancement of natural killer T lymphocyte plasticity in murine models of immune-mediated disorders. Gut. 2007;56:82–89. doi: 10.1136/gut.2006.095497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, analytic methods, and study materials have been made available in the results, figures, and materials and methods of this article.