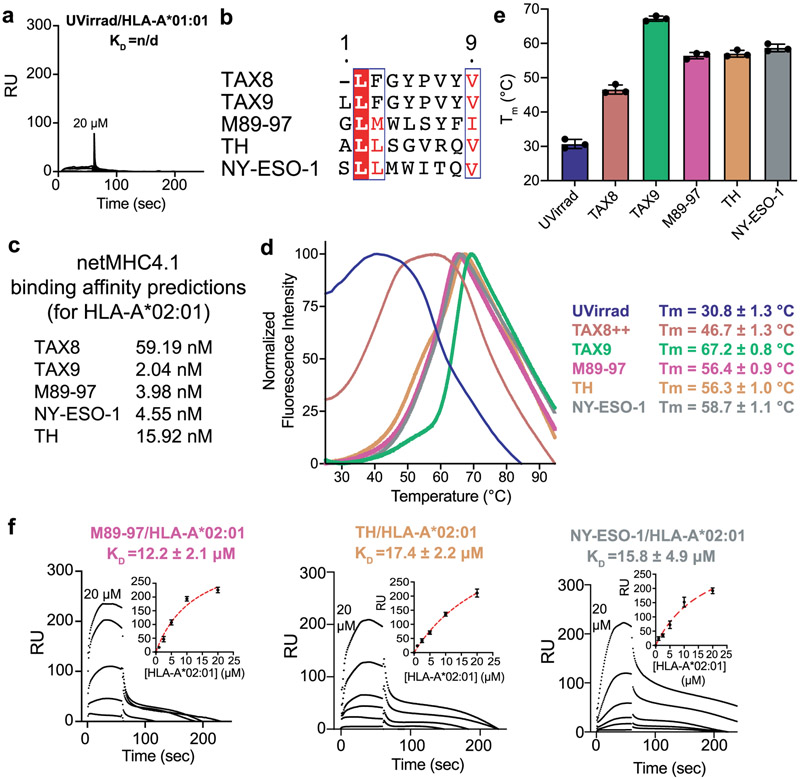
Extended Data Fig. 5 ∣. Constructs for interaction of properly conformed MHC-I with TAPBPR.
a, Representative SPR sensorgram for varying concentrations of UV-irradiated HLA-A*01:01/hβ2m flowed over a streptavidin chip coupled with TAPBPRWT-biotin. RU = response units. b, Clustal Omega v1.2.4 alignment of HLA-A*02:01 peptides used in this study, processed using ESPript v3. Residues in red box, white character represent strict identity. Residues in red character represent similar group amino acids. c, In silico prediction of peptide binding to HLA-A*02:01 using netMHCpan-v4.1. d, Normalized DSF traces of purified empty and peptide-loaded loaded HLA-A*02:01/hβ2m. Fitted thermal stabilities (Tm, °C) are noted. Data are mean ± SD for n = 3 technical replicates. e, Summary of DSF determined thermal stability (Tm, °C) of empty (UVirrad) or peptide-loaded HLA-A*02:01/hβ2m complexes. Data are mean ± SD for n = 3 technical replicates. f, Representative SPR sensorgrams for varying concentrations of non-UV irradiated M89-97/HLA-A*02:01/hβ2m, TH/HLA-A*02:01/hβ2m, or NY-ESO-1/HLA-A*02:01/hβ2m flowed over a streptavidin chip coupled with TAPBPRWT-biotin. The concentration of analyte for the top sensorgram is noted. Fits from surface bound analysis are shown with dotted red lines. Data are mean ± SD for n = 3 technical replicates.