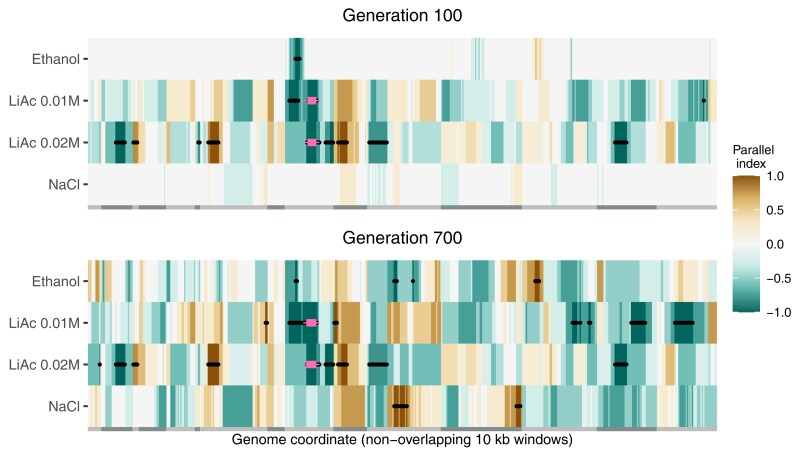
Fig. 5.
Heatmaps of genetic parallelism along the genome in the different environments. The genome was divided into non-overlapping 10 kb windows with at least four SNPs and the median allele frequencies were used to calculate a parallelism index. The index is a scaled count of how many replicates are fixed (MAF ≤ 0.1) for the SK1 parental allele (+1) or the Y55 parental allele (−1). Chromosome limits are defined by alternating light and dark gray bars at the bottom of the heatmaps. Black points mark windows where either parental allele was fixed in four or more replicates (LiAc 0.02 M has five replicates, all other environments had four). Pink crosses mark windows that were completely fixed in all the LiAc replicates (both LiAc 0.01 M and LiAc 0.02 M).