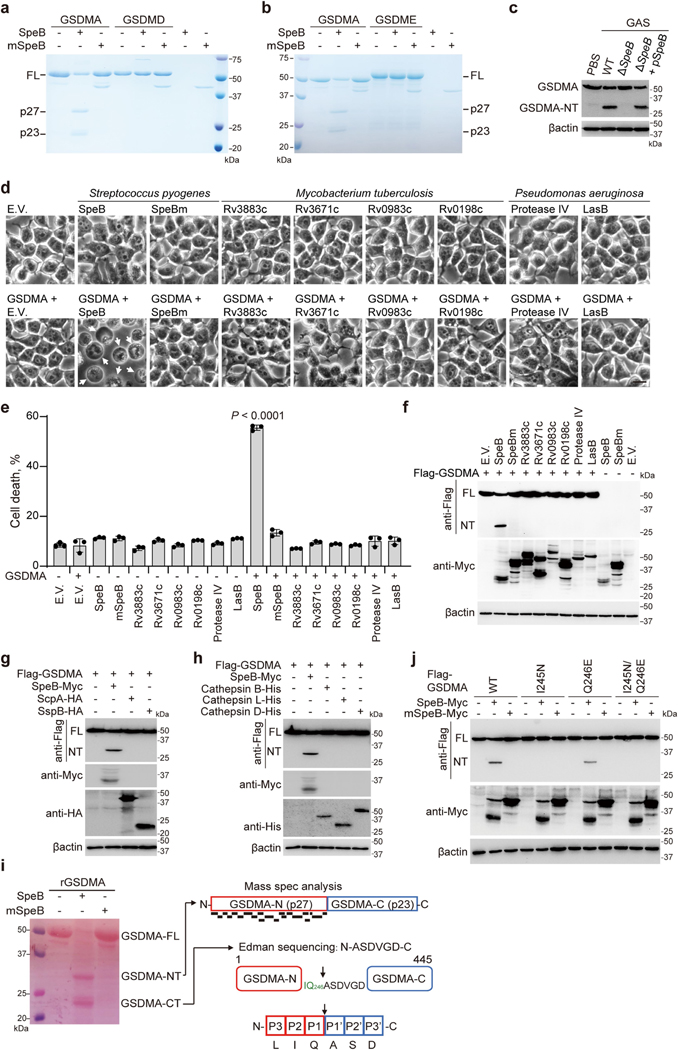
Extended Data Fig. 5 |. SpeB specifically targets and cleaves GSDMA.
a, b, In vitro cleavage assay of recombinant GSDMA, GSDMD (a) or GSDME (b) by incubation with recombinant WT SpeB (100 nM) or mSpeB (250 nM) for 0.5 h. c, Whole cell lysates of A431 cells infected or not with the indicated GAS strains were subjected to immunoblot analysis for the indicated proteins. d–f, 293T cells were transfected with the indicated plasmids (Flag-tagged GSDMA, Myc-tagged bacterial proteases) before analysed by phase-contrast microscopy (d), LDH release (e) and immunoblot of whole cell lysates with the indicated antibodies (f). g, h, Flag-tagged GSDMA was treated or not with SpeB, staphopains (ScpA, SspB), or cathepsins (Cathepsin B, L, D) before subjected to immunoblot analysis with the indicated antibodies. i, GSDMA N-terminal and C-terminal cleavage products (p27 and p23) were analysed by mass spectrometry (MS) and Edman sequencing, respectively. Individual peptides identified by MS and shown in black bars were mapped against N-terminal GSDMA (upper right panel). Middle right panel shows the N-terminal sequence of p23 determined by Edman sequencing, the diagram of GSDMA two-domain architecture, and SpeB cleavage site Gln246 highlighted in green. Bottom panel shows the positions (P) on the substrate of SpeB (GSDMA) which are counted and numbered (P3-P2-P1-P1’-P2’-P3’) from the point of cleavage. j, Whole cell lysates of 293T cells transfected with the indicated plasmids were collected and subjected to immunoblot analysis. Scale bar: 10 μm. Data are representative of at least three independent experiments. Graphs show mean ± s.d. of triplicate wells. e, One-way ANOVA. Data are representative of at least three independent experiments. For gel source data, see Supplementary Fig. 1.