Figure 1.
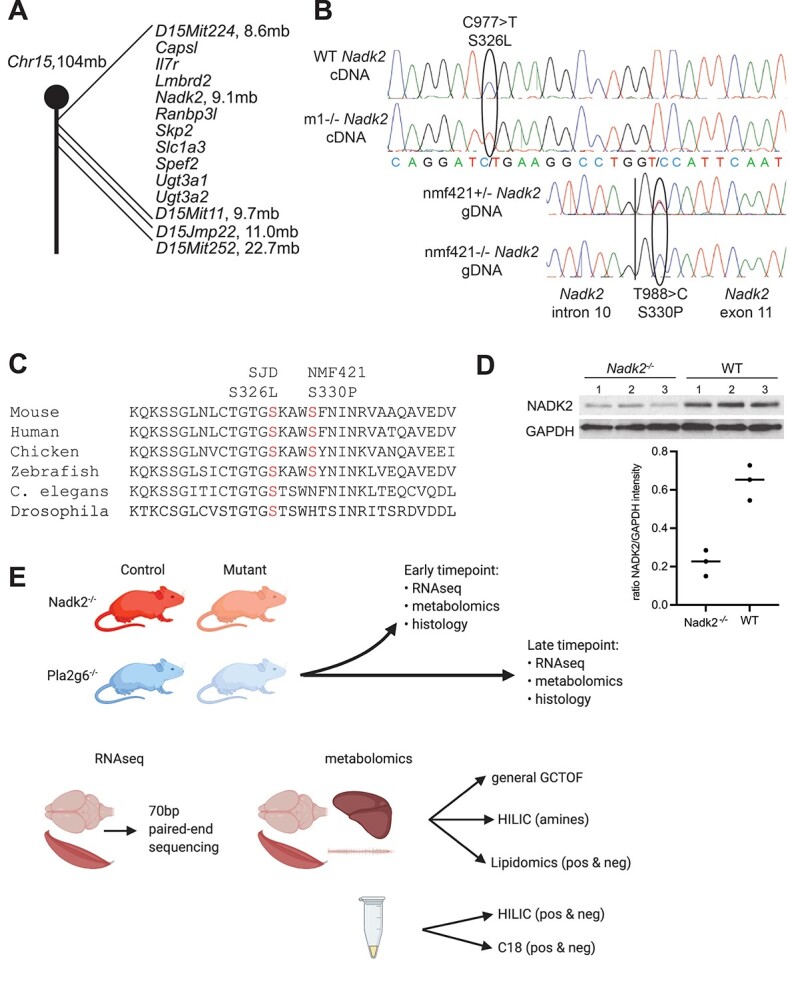
Nadk2 mouse mutations and study design. (A) The ENU-induced mutations mapped to proximal mouse Chromosome 15, with the m1Jcs allele mapping between D15Mit224 at 8.6 mb and D15Mit11 at 9.7 mb. This interval contains 10 protein coding genes. The nmf421 allele mapped proximally to D15Jmp22 at 11.0 mb. (B) Sequencing chromatograms showing the C977 > T mutation in cDNA prepared from wild type and m1Jcs homozygous mice (m1), and the T988 > C mutation in PCR-amplified genomic DNA from heterozygous and homozygous nmf421 mice. The vertical line in genomic DNA sequence is the intron 10/exon 11 boundary. (C) Evolutionary conservation of S326 and S330 in NADK2. (D) Western blot analysis of NADK2 versus a GAPDH loading control revealed the mutant protein were significantly reduced compared to littermate controls. (E) Overview of RNA-Seq and metabolomics studies. Samples from Nadk2 and Pla2g6 mutant mice, as well as wild-type littermates for each, were collected at both early and late time points as described in the ‘Materials and Methods’. At each time point, samples were collected for RNA-Seq and metabolomics, as well as for histology. Samples for RNA-Seq derived from whole brain and muscle, and were sequenced using 100-bp paired-end sequencing. Samples for metabolomics included whole brain, muscle, spinal cord, and liver, as well as plasma, which was previously omitted. These samples were analyzed by general GCTOF mass spectrometry, HILIC for biogenic amines, and lipidomics (positive and negative ionization modes).
