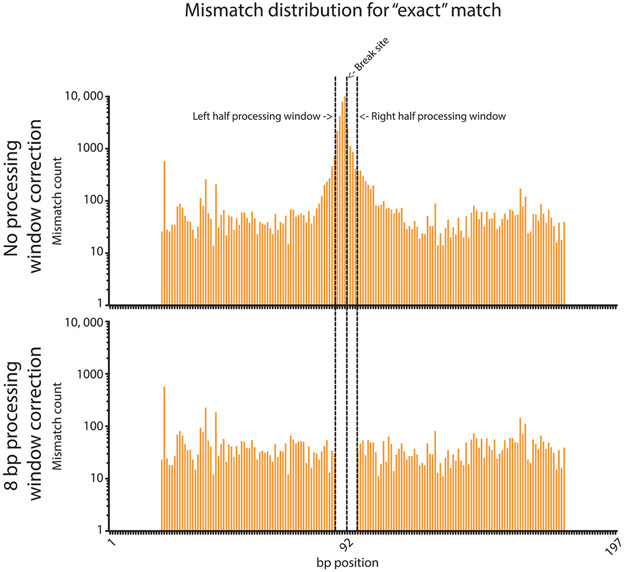
Fig. 3.
Distribution of base substitutions in Illumina sequencing DSB repair amplicons. The distribution of mismatched bp is given for reads that were considered perfect repair events by Geneious, before and after the 8 bp search for base substitutions correction. The majority of mismatches occur homogenously across the read, as expected of sequencing error, with counts of ~10–100. However, the mismatches peak around the break site with a maximum of 11,982 immediately at the break site (~120–1200-fold higher than the surrounding sequence). The 8 bp processing window correction recategorizes these counts and leaves the rest that are likely sequencing error. No mismatches occur at the ends of the reads due to the “Trim_and_Pad” script used to filter reads.