Figure 2. Aging abolishes rhythmic gene expression in macrophages.
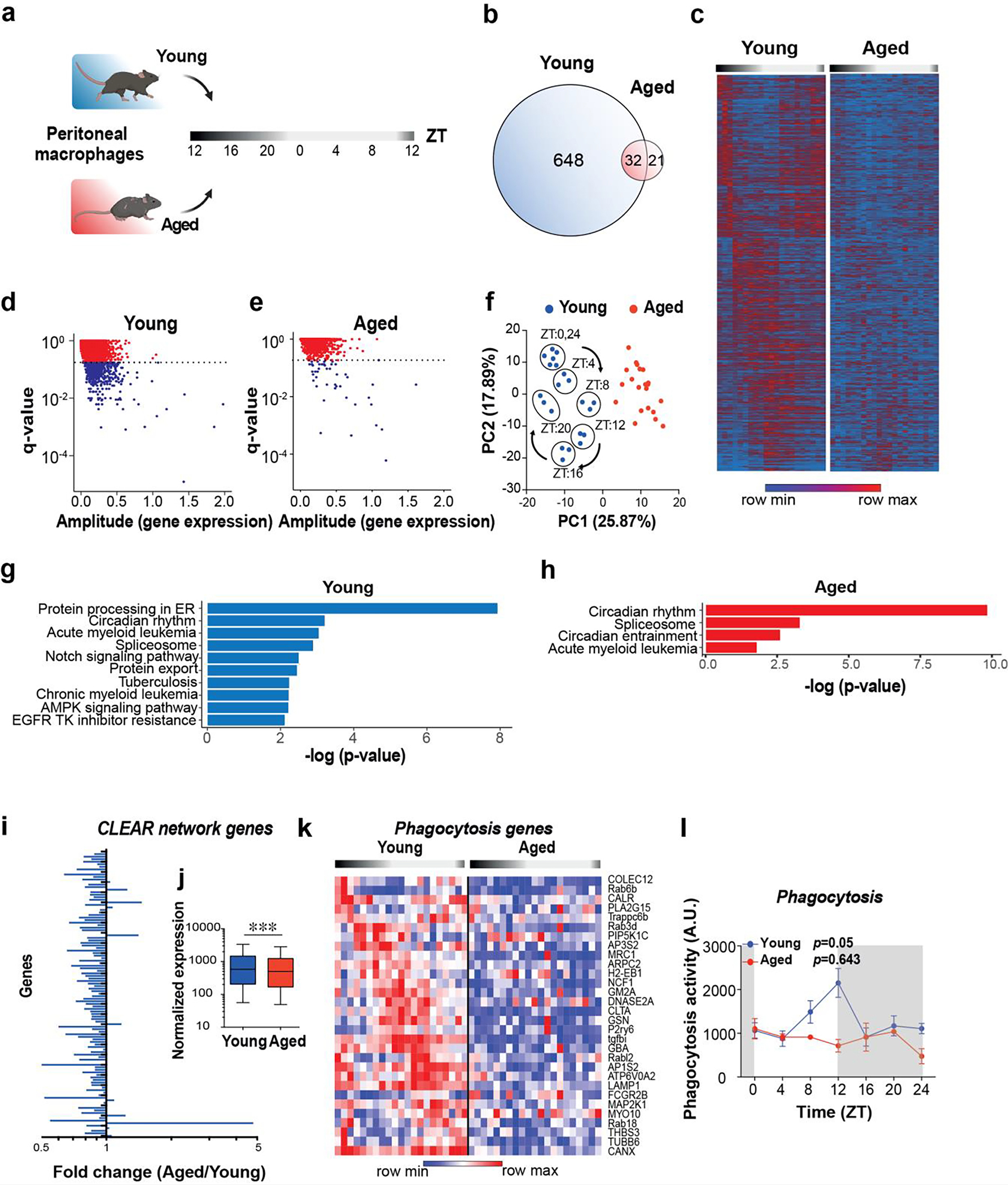
a. Schematic of experimental design. Peritoneal macrophages were collected from young (2 mo) and aged (20–22 mo) male mice at 4-hour intervals over a 24-hour period for RNA-seq; n=21 mice in each age group and n=3 in each time interval.
b. Venn diagram of unique and shared rhythmically expressed transcripts in young vs. aged macrophages.
c-e. Heatmap (c) and scatterplots of JTK_CYCLE results (d-e) of diurnally oscillating transcripts in young and aged peritoneal macrophages. The dotted lines in d and e represent q-value cutoffs of 0.2.
f. Principal component analysis (PCA) of rhythmic transcripts from young and aged macrophages. Note circadian clustering observed in young mice that is absent in the aged group.
g-h. KEGG pathway analyses of transcripts that show diurnal oscillations in young (g) and aged (h) macrophages. Genes with p<0.05 and q<0.1 by JTK_CYCLE are shown.
i. Fold change of Coordinated Lysosomal Expression and Regulation (CLEAR) network gene expression levels in aged vs young peritoneal macrophages over all time points.
j. Pooled normalized expression levels of CLEAR network transcripts in young and aged peritoneal macrophages; p < 0.001, 2-sided Mann Whitney U test.
k. A heatmap showing genes belonging to the KEGG pathway of phagocytosis over a 24 h time period in young and aged peritoneal macrophages. Each column represents one mouse. (n=3 mice per age per time interval). q<0.2, JTK_CYCLE.
l. Young and aged peritoneal macrophages were assayed for phagocytosis of fluorescent E coli particles over a 24 hour period, p=0.05 and p=0.643 for young and aged, respectively, by JTK_CYCLE (n=3 mice per time point per age).
