Figure 2.
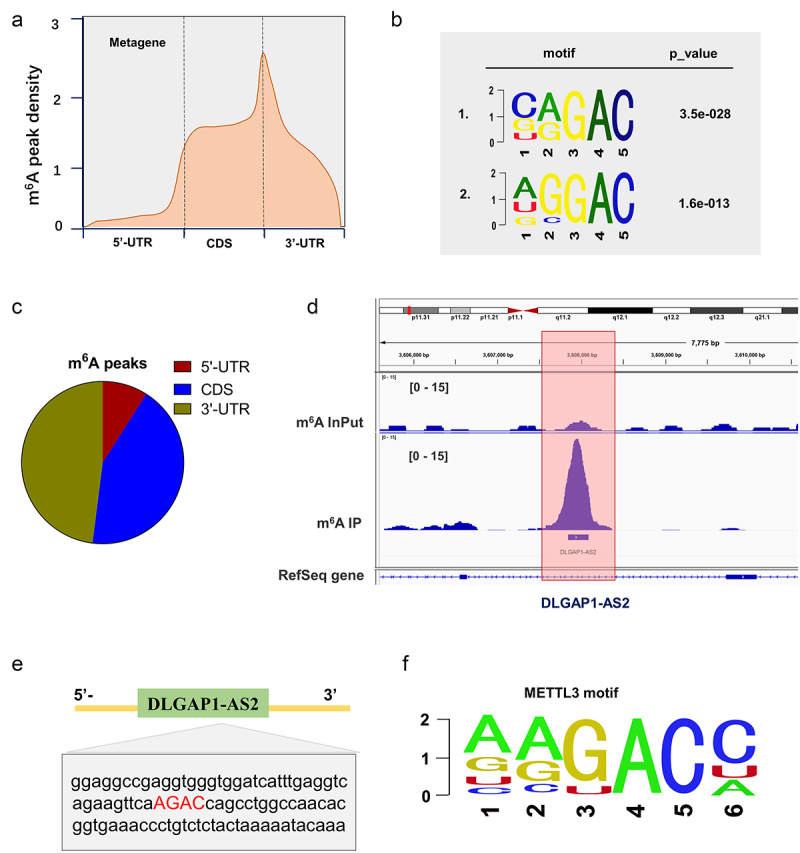
MeRIP-Seq revealed the m6A modification of DLGAP1-AS2. (a, c) MeRIP-Seq revealed the m6A peaks density in NSCLC cells, including 5’-UTR, coding region (CDS) and 3’-UTR. (b) The significant m6A motif for the m6A peaks was GGAC. (d) MeRIP-Seq showed that there was a remarkable m6A modification site in the 3’-UTR of DLGAP1-AS2. (e) Genomic analysis displayed the sequences of DLGAP1-AS2 3’-UTR containing m6A site (AGAC). (f) The motif of METTL3 targeting DLGAP1-AS2 m6A site.
