Figure 1.
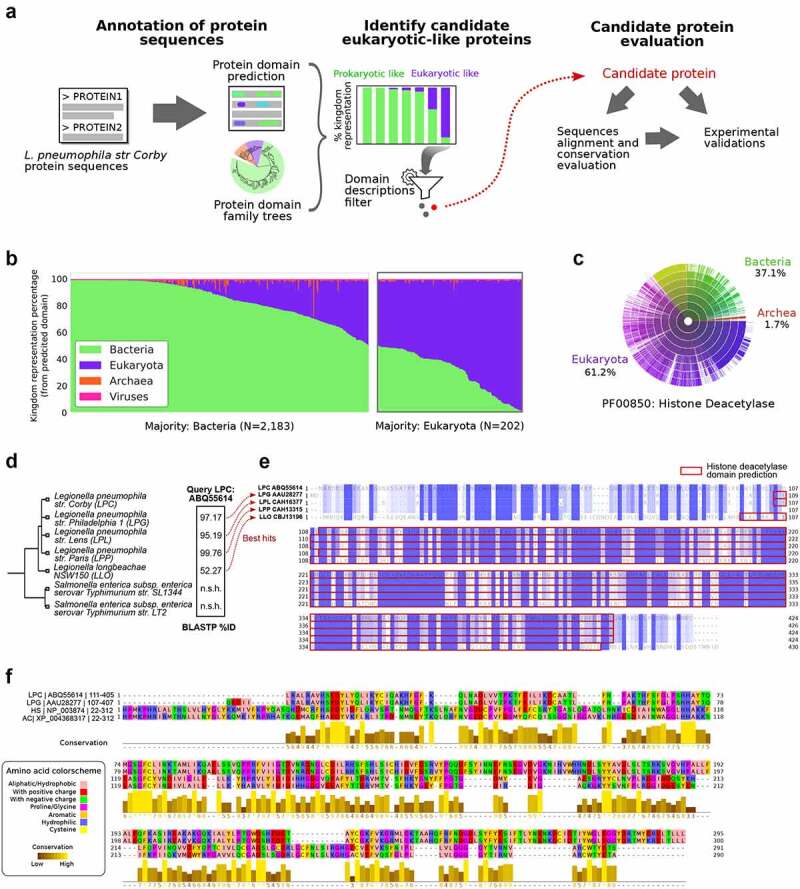
Screening of candidate eukaryotic-like proteins in L.P. strain Corby identifies Smh1 as a candidate histone-deacetylase. (a) Pipeline of analyses from the in silico annotation of protein sequences of L.P. strain Corby to the identification of candidate proteins from their eukaryotic-like protein domains, screened for enzymatic functions suggesting potential impact on the host regulatory machinery. (b) a total of 2,385 proteins were annotated with protein domains from Pfam database. Relative representation of annotated protein domain across the different kingdoms of life highlights a subset of 202 proteins bearing domains mostly represented in eukaryotic sequences. (c) Representation across kingdoms of the Pfam domain PF00850 “histone-deacetylase,” annotated in the sequence of the candidate protein ABQ55614 from L.P. strain Corby. (d) Results of the BLASTP search for homologous proteins to ABQ55614 in L.P. subspecies as well as Salmonella species. Percentage of sequence identity are reported for best hits; “n.S.h:” no significant hit. (e) Clustal omega multiple sequence alignment of the orthologs of ABQ55614 identified in L.P. subspecies. Blue colouring indicates conservation levels, while red frames indicate the independent predictions of the histone-deacetylase domains in each protein sequence. (f) Jalview visualization of the clustal omega alignment of histone-deacetylase domains extracted from L.P. proteins from strains Corby and Philadelphia, as well as from the Human Histone-deacetylase 3 (HDAC3, NCBI protein ID NP_003874) and the protein “XP_004368317” from A. castellanii. Amino acids are coloured to highlight physicochemical properties (Zappo colour scheme). Yellow barplots identify levels of conservation.
