Figure 2.
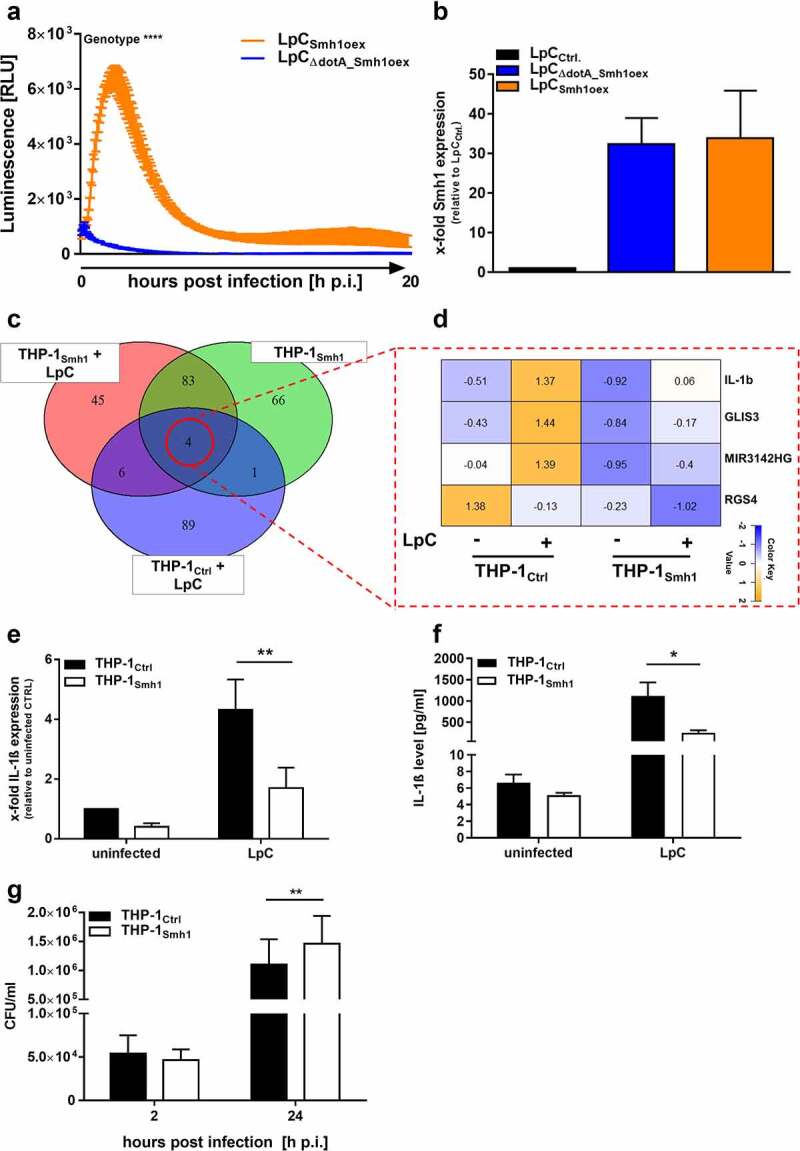
Effector protein Smh1 translocates into host cytosol and influences gene expression and Legionella pneumophila replication. (a) RAW 264.7 cells expressing cytosolic LgBit were infected with L.P. (T4SS-capable as well as the T4SS-incapable ΔdotA mutant) expressing HiBit-tagged Smh1 at MOI 50. Luminescence was detected over a period of 20 h. Two-way ANOVA with Sidak´s correction was performed and data are shown as mean ± SEM of at least three independent biological replicates. ****p ≤ 0.0001 for comparison between background genotypes. (b) Overexpression of Smh1 in LpC was verified by qPCR. (C-F) THP-1 cells expressing Smh1 (THP-1Smh1) and corresponding control cells (THP-1Ctrl) were stimulated with PMA (80 nM) for 72 h and infected with L. pneumophila Corby (LpC) at MOI 1 for 3 h (C-E) or 24 h (F) or left uninfected. (c) Venn representation shows significantly (padj <0.05) regulated genes after expression of Smh1 alone or additional LpC infection in comparison to infected Ctrl. Expression data for every section is provided in Table S1. (d) the four genes from the central Venn overlap are shown with their z-scores computed on normalized read counts from DeSeq2. (e) IL-1β expression was examined by qPCR and is shown as fold change of the uninfected Ctrl. (f) IL1β release was measured by ELISA. (g) PMA-differentiated THP-1 cells were infected with LpC at MOI 10. Bacterial replication was analysed by colony forming unit (CFU) Assay 2 and 24 hours post infection (h p.I.). (E-G) Two-way ANOVA with Sidak´s correction was performed and data are shown as mean + SEM of at least three independent biological replicates. *p ≤ 0.05; **p ≤ 0.01 (compared to infected Ctrl).
