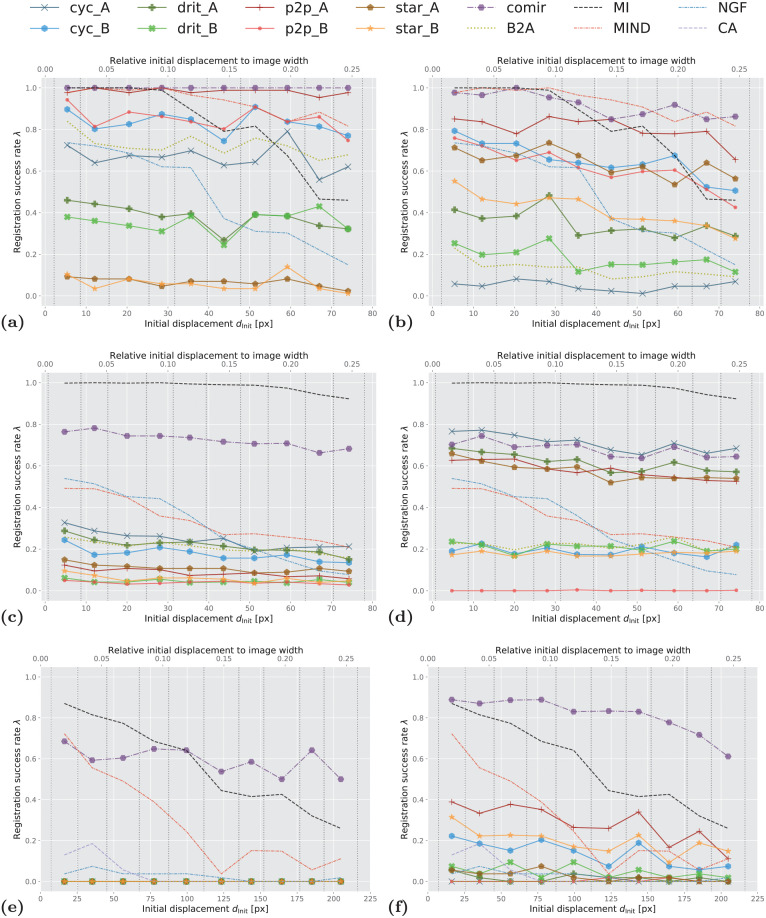
Fig 8. Success rate of the observed registration approaches.
(A) SIFT on Zurich data, (B) α-AMD on Zurich data, (C) SIFT on Cytological data, (D) α-AMD on Cytological data, (E) SIFT on Histological data, (F) α-AMD on Histological data. x-axis: initial displacement dInit between moving and fixed images, discretised into 10 equally sized bins (marked by vertical dotted lines). y-axis: success rate λ within each bin (averaged over 3 folds for Zurich and Cytological data). In the legend, cyc, drit, p2p, star and comir denote CycleGAN, DRIT++, pix2pix, StarGANv2, and CoMIR methods respectively. Suffix _A (resp. _B) denotes that generated Modality A (resp. B) is used for the (monomodal) registration. B2A denotes registration of the original multimodal images, without using any modality translation. MI, MIND, NGF and CA represent using MI maximisation, MIND, NGF and CurveAlign for registration, respectively.