Fig. 5. Adoptively transferred Ikaros-silenced BMMs attenuate liver IRI in CD11b-DTR mice by upregulating SIRT1 and suppressing caspase-1-GSDMD processing.
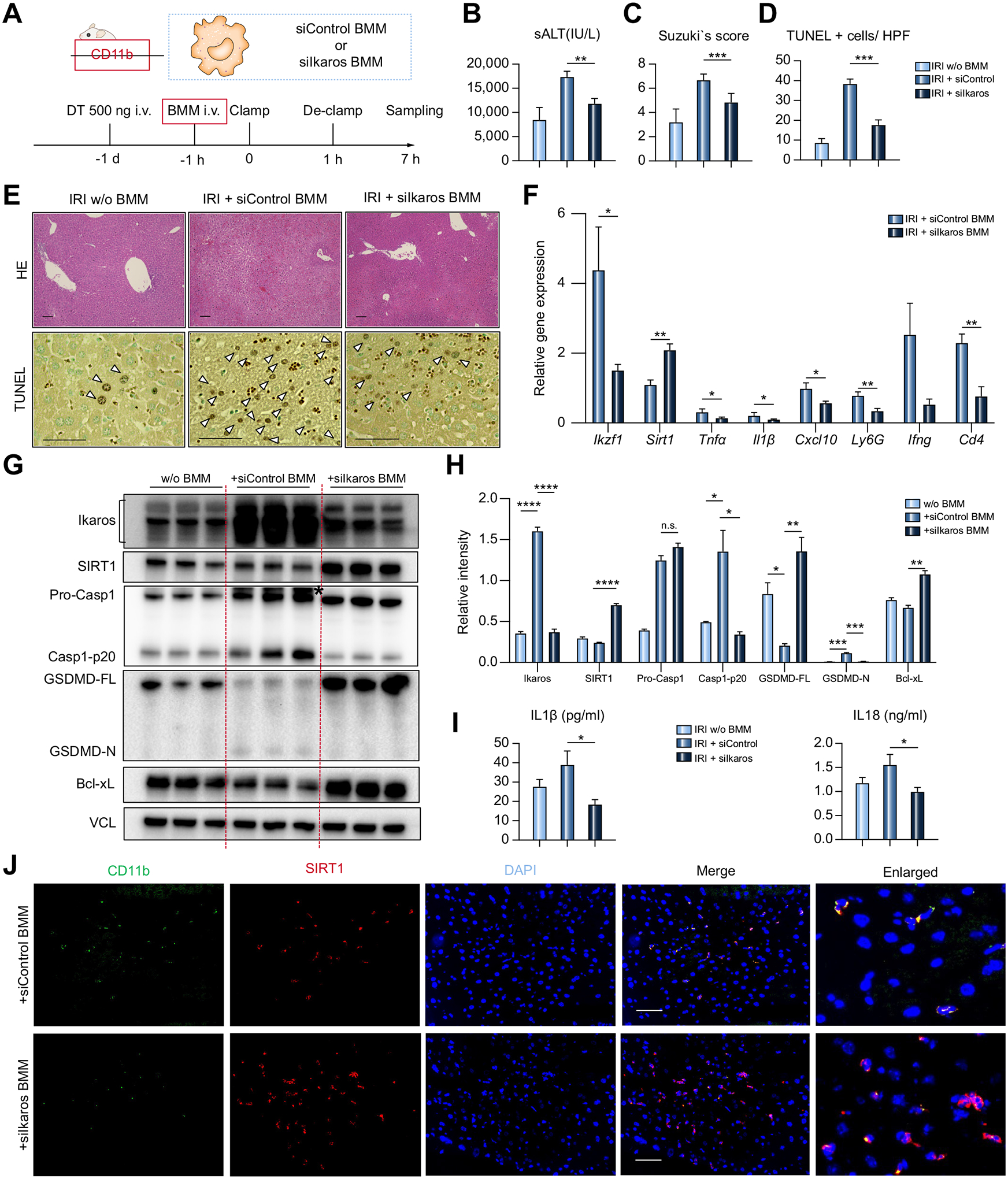
CD11b-DTR mice untreated or reconstituted with BMMs transfected with siControl vs. siIkaros RNAs were subjected to 60 min warm ischemia and 6 h reperfusion. (A) Workflow of CD11b+ cell depletion/reconstitution by BMMs, followed by warm liver IRI. (B) sALT levels (n = 5–6/group). (C) Suzukìs histological grading of liver IRI and (D) quantification of TUNEL+ cells/HPF (n = 5–6/group). (E) Representative H&E (original magnification, ×100; scale bar, 100 μm) and TUNEL (original magnification, ×400; scale bar, 50 μm) staining. Arrowheads show TUNEL+ cells. (F) qRT-PCR-assisted detection of mRNA coding for Ikzf1, Sirt1, Tnfα, Il1β, Cxcl10, Ly6G, Ifnγ, and Cd4. Data were normalized to Gapdh expression (n = 3–4/group). (G) Western blot-assisted detection of Ikaros, SIRT1, Pro-Casp1, Casp1-p20, GSDMD, Bcl-xL, and VCL. Asterisk indicates non-specific band. (H) Relative intensity ratios with VCL normalization (n = 3/group). (I) Serum IL1β/IL18 levels (n = 5–6/group). (J) Representative immunohistochemical images illustrate hepatic detection of CD11b (green), SIRT1 (red) in CD11b-DTR mice repopulated with siControl or siIkaros BMM (original magnification, ×400, scale bar, 50 μm). Data shown are mean ± SEM. *p <0.05; **p <0.01; ***p <0.001; ****p <0.0001 by Student’s t test. BMM, bone marrow-derived macrophages; HPF, high power field; IRI, ischemia-reperfusion injury; LPS, lipopolysaccharide; qRT-PCR, quantitative reverse-transcription PCR; sALT, serum alanine aminotransferase; si(RNA), small-interfering RNA.
