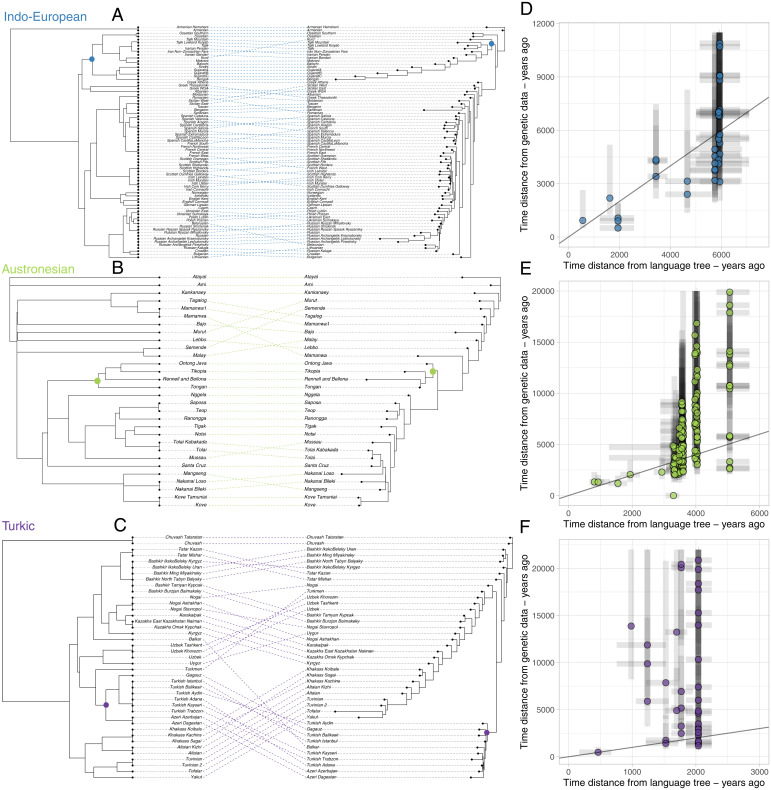
Fig. 4.
Genetic and linguistic relatedness within three language families: Indo-European, Austronesian, and Turkic. (A–C) Comparison between a linguistic tree (Right) generated with Bayesian models of cognate replacement (34, 48, 49) and a genetic tree generated with FST distances (Left) for each language family. Each taxon corresponds to a population of the database; in some cases, the same language is spoken by different populations. In the trees, the colored dots mark clades which include the same populations in both trees: Indo-Aryan in Indo-European, Polynesian in Austronesian, and Nuclear Oghuz in Turkic. (D–F) correlation between linguistic and genetic divergence time (for the pairs for which it is possible to reconstruct effective population size). Gray bars mark 95% credible intervals from linguistic trees and 5 to 95 percentile intervals associated with the genetic divergence time reconstructions. The black line marks a 1:1 correspondence. Genetic outliers have been excluded, namely, Sardinian, Rennell and Bellona, and Mamanwa, which return very ancient divergence times (possibly due to drift and/or ancient admixture).