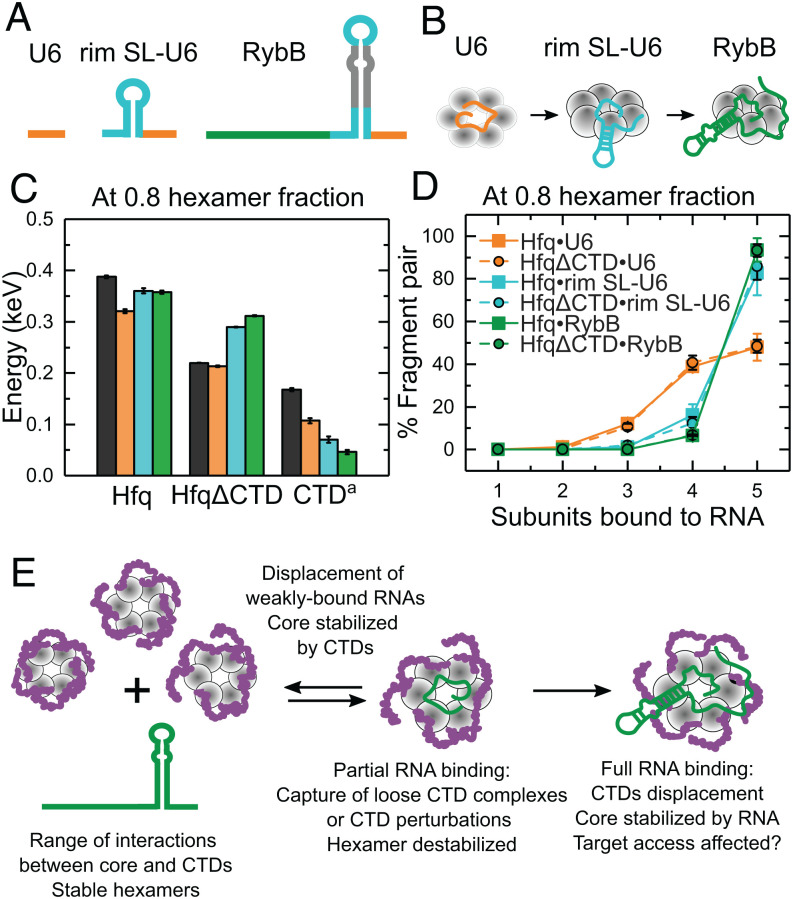
Fig. 5.
RNA binding progressively displaces the Hfq CTDs. (A) RNAs mimicking segments of RybB sRNA that progressively interact with more Hfq surfaces: 3′ U tail (orange), terminal stem-loop (cyan), 5′ seed (dark green). (B) Cartoon of progressive RNA binding. (C) Stabilities of Hfq, HfqΔCTD (core) and conferred by the CTDs a(Hfq•RNA-HfqΔCTD•RNA) when bound to the RNAs shown in A. Colors correspond to the RNAs in B. Black bar, protein only. Stabilities and errors as in Fig. 4C (SI Appendix, SI Materials and Methods). (D) Percentage of fragment pairs after the collision of Hfq (solid lines, squares) or HfqΔCTD (dashed lines, filled circles) bound to the RNAs in A (see also SI Appendix, Fig. S13). Errors as in Figs. 2 D and E. (E) Assorted organization of the disordered CTDs results in heterogeneous hexamers. RNAs could bind hexamers with open surfaces or perturb a CTD. Local CTD disturbances destabilize the hexamer and are transmitted to the rest of the protein, promoting further CTD displacement and stable binding for favorable RNAs. Nonpreferred RNAs are displaced by competition with the CTDs (11), reestablishing hexamer stability.