Fig. 7. Dapl1-mediated NFATc2 inhibition contributes to regulation of Havcr2 expression and CD8 T cell functions.
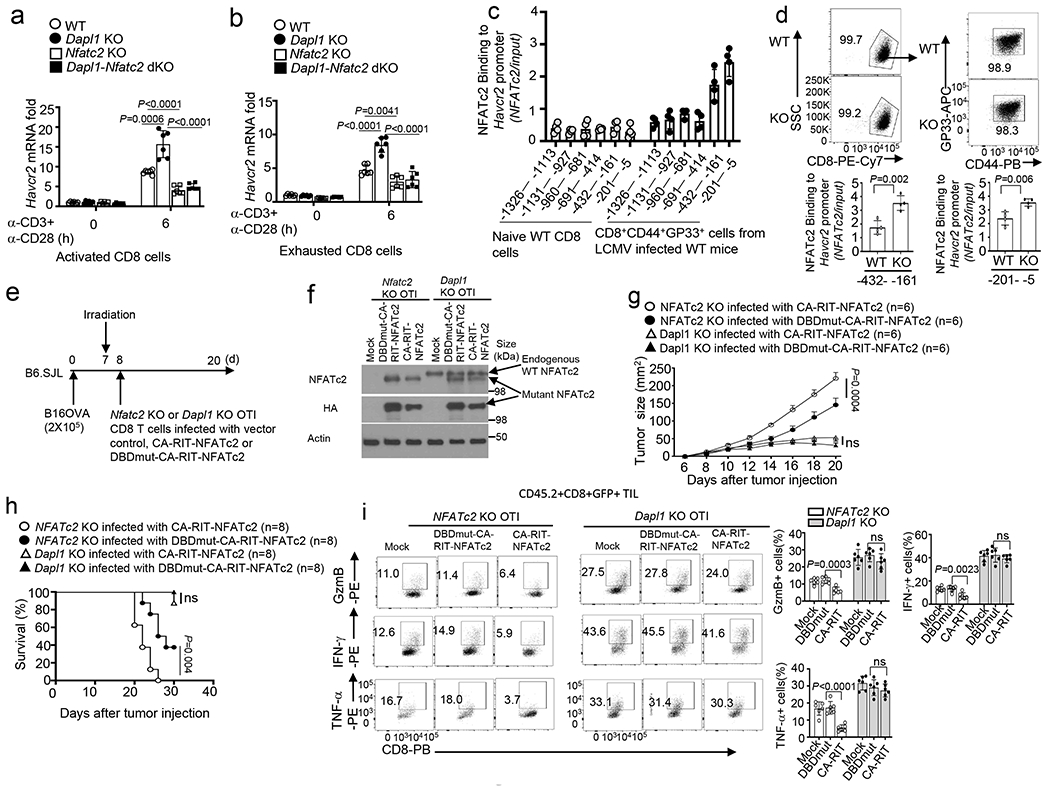
a,b, qRT-PCR analysis of Havcr2 mRNA in activated CD8 T cells (a) or in vitro exhausted CD8 T cells (b) from the indicated mouse strains, stimulated for 6 h with anti-CD3 and anti-CD28. n =6 per genotype. c, ChIP assay of the recruitment of NFATc2 to various regions of the Havcr2 promoter in naive CD8 T cells from uninfected wildtype mice or CD8+CD44+GP33+ cells from LCMV clone 13-infected (for 4 weeks) mice. n =4 per genotype. d, ChIP assays of the binding of NFATc2 to the indicated regions of Havcr2 promoter in CD8+CD44+GP33+ cells derived from wildtype or Dapl1 KO mice infected with LCMV clone 13 for 4 weeks. n =4 per genotype. e,f, Schematic of experimental design (e) and immunoblot analysis of endogenous wildtype NFATc2 and transduced HA-tagged NFATc2 mutants in Nfatc2 KO or Dapl1 KO CD8 T cells infected with vector control (Mock), CA-RIT-NFATc2 or DBDmut-CA-RIT-NFATc2 (f), n = 2 biologically independent experiments. g-i,Tumor growth curve (g), mouse survival plot (h), and flow cytometry analysis of the frequency of IFN-γ-, TNF-α- and Granzyme B-producing CD8 T cells in the tumor (i) of B16-OVA tumor-bearing B6.SJL mice adoptively transferred with the indicated OTI effector CD8 T cells depicted in e. n = 6 (g,i) or n = 8 (h) per genotype. Data are pooled from two independent experiments (e-i), Summary data are shown as the mean±s.d. with P values determined using a two-tailed unpaired Student’s t-test (a,b,d,i), two-sided log-rank Mantel–Cox tests (h) and two-way ANOVA with Bonferroni correction (g). ns, not significant.
