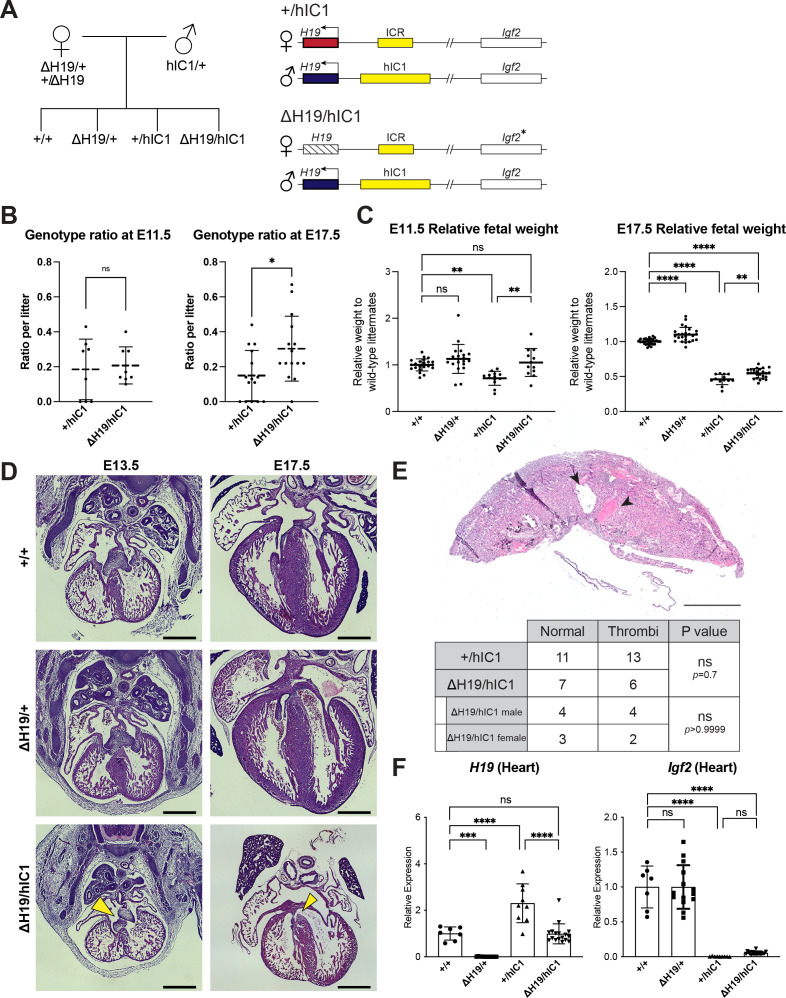
Figure 4. Normalizing H19 expression through the maternal deletion of H19.
(A) A schematic representation of the rescue breeding between △H19 heterozygous female and hIC1/+ male mice. △H19/hIC1 embryos are expected to express H19 only from the paternal allele and maternally express Igf2 in a tissue-specific manner. (B) Ratio of +/hIC1 and △H19/hIC1 embryos observed in E11.5 and E17.5 litters (mean ± SD). 8 E11.5 litters and 15 E17.5 litters with litter size larger than five pups were examined. Each data point represents one litter. (C) Relative fetal weights of wild-type, △H19/+, +/hIC1, and △H19/hIC1 embryos at E11.5 and E17.5, normalized to the average body weight of the wild-type littermates (mean ± SD). (D) Representative cross-sections of wild-type, △H19/+ and △H19/hIC1 embryonic hearts at E13.5 and E17.5, stained with hematoxylin and eosin. Note the cushion defect at E13.5 and the ventricular septal defect (VSD) at E17.5 in △H19/hIC1 hearts (yellow arrows). All E13.5 samples and E17.5 wild-type sample are male, and E17.5 △H19/+ and △H19/hIC1 samples are female. Scale bars = 500 µm. (E) Top: Representative cross-section of E17.5 △H19/hIC1 male placenta stained with hematoxylin and eosin. Black arrowheads indicate thrombi. Scale bar = 1 mm. Bottom: Number of the wild-type, male, and female +/hIC1 placentas with thrombi observed. (F) Relative total expression of H19 and Igf2 in E17.5 wild-type, △H19/+, +/hIC1, and △H19/hIC1 hearts (mean ± SD). (C and F) Each data point represents an individual conceptus from different litters. Statistics used are (B, C, and F) one-way ANOVA with Tukey’s multiple comparisons test and (E) Fisher’s exact test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, and ns = not significant.