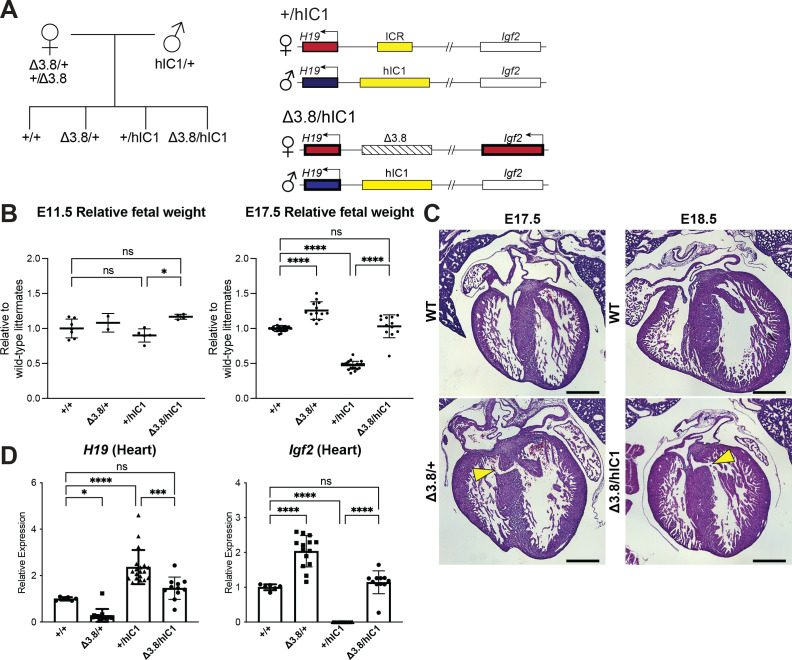
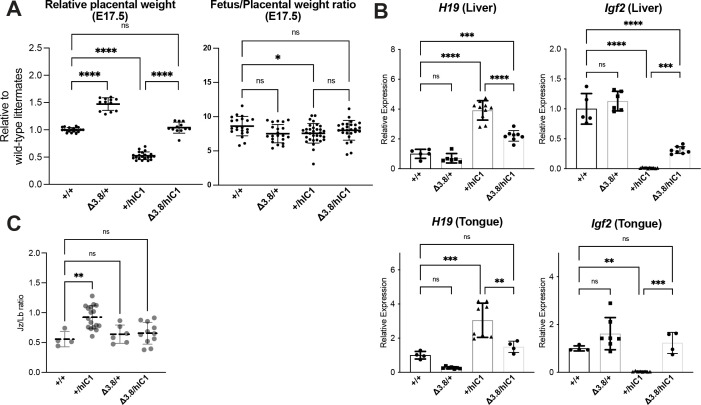
Figure 5. Restoring H19 and Igf2 expression utilizing maternal H19/Igf2 imprinting control region (ICR) deletion.
(A) A schematic representation of the offspring produced when △3.8 heterozygous female and hIC1/+ male mice are mated is depicted. △3.8/hIC1 embryos are expected to show activation of maternal Igf2 expression as well as paternal H19 expression. (B) Relative fetal weights of wild-type, △3.8/+, +/hIC1, and △3.8/hIC1 embryos at E11.5 and E17.5, normalized to the average body weight of wild-type littermates (mean ± SD). 2 E11.5 litters and 10 E17.5 litters are presented. (C) Representative cross-sections of E17.5 △3.8/+ and E18.5 △3.8/hIC1 embryonic hearts with ventricular septal defects (VSDs; yellow arrows), stained with hematoxylin and eosin. Sections from wild-type littermates are shown together for comparison. The E17.5 wild-type sample is male, and the rest are female. Scale bars = 500 µm. (D) Relative total expression of H19 and Igf2 in E17.5 wild-type, △3.8/+, +/hIC1, and △3.8/hIC1 hearts (mean ± SD). (B and D) Each data point represents an individual conceptus from different litters. One-way ANOVA with Tukey’s multiple comparisons test was used with *p<0.05, ***p<0.001, ****p<0.0001, and ns = not significant.