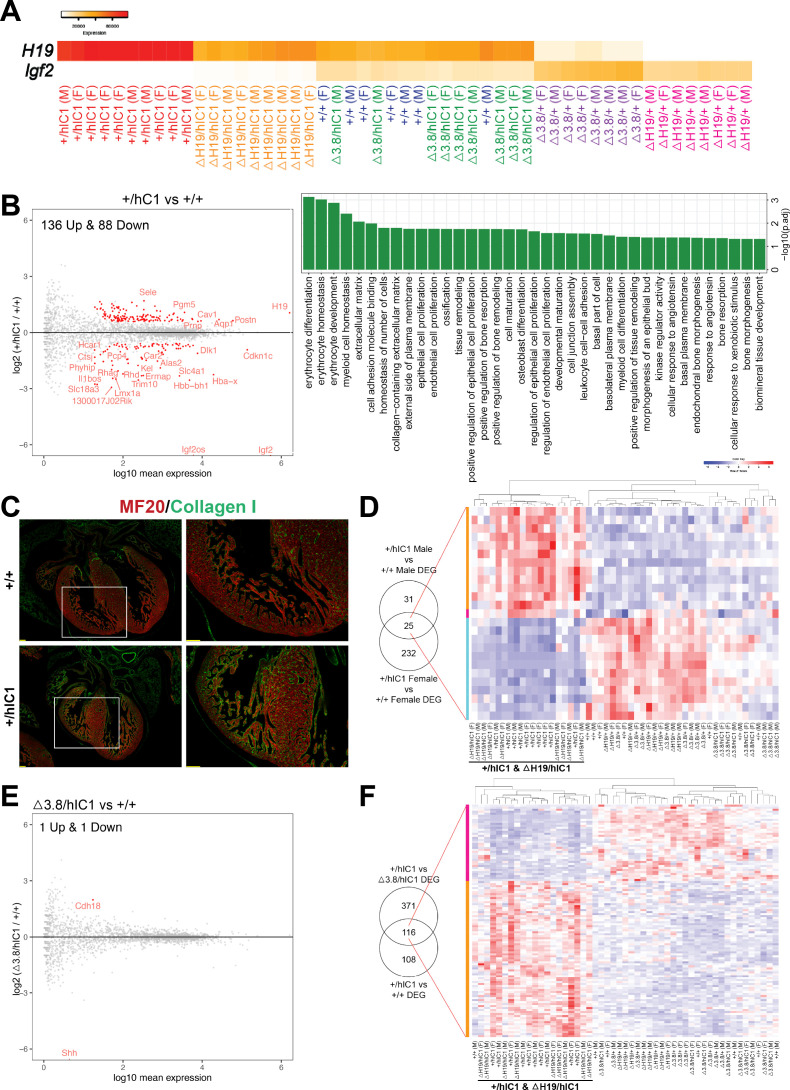
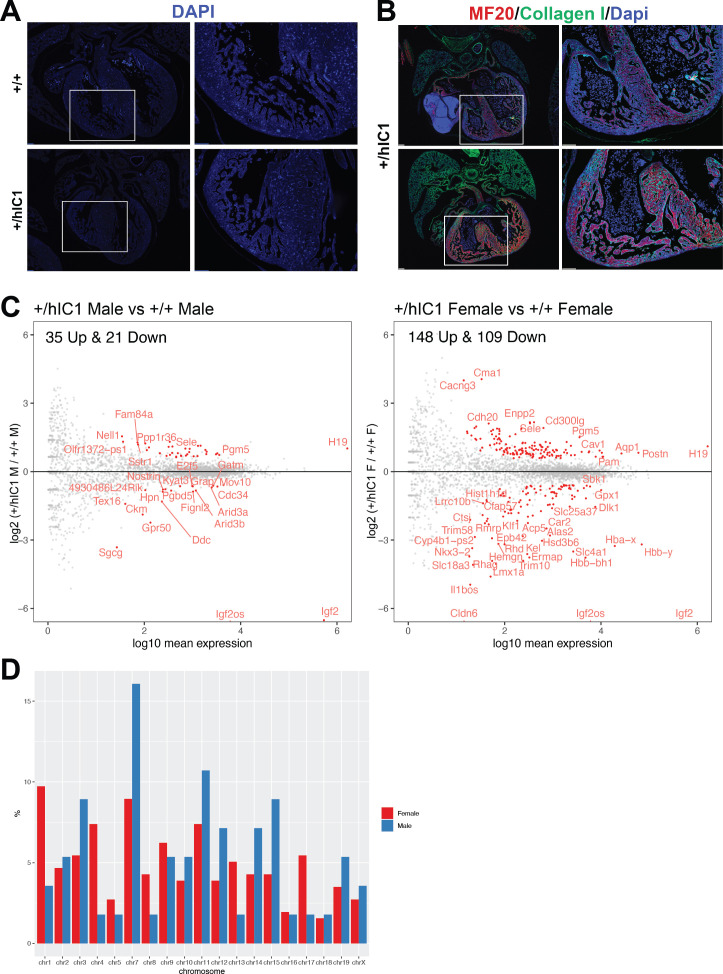
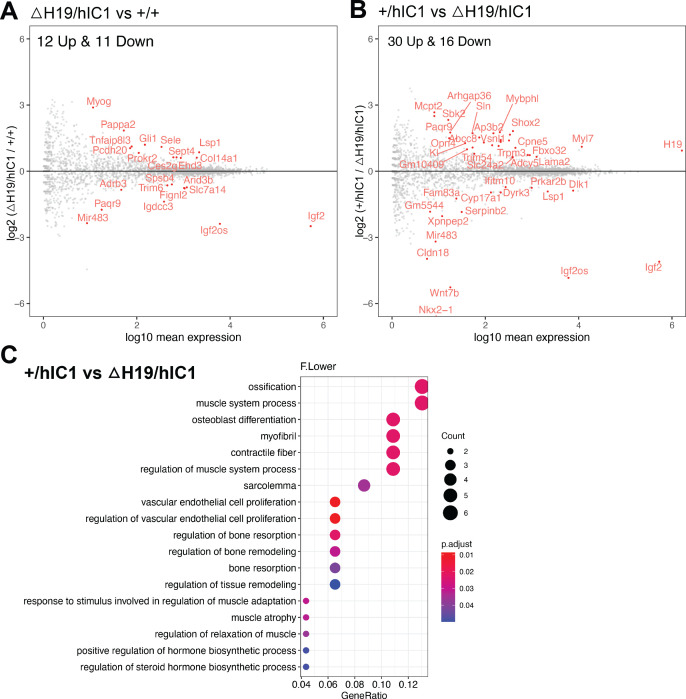
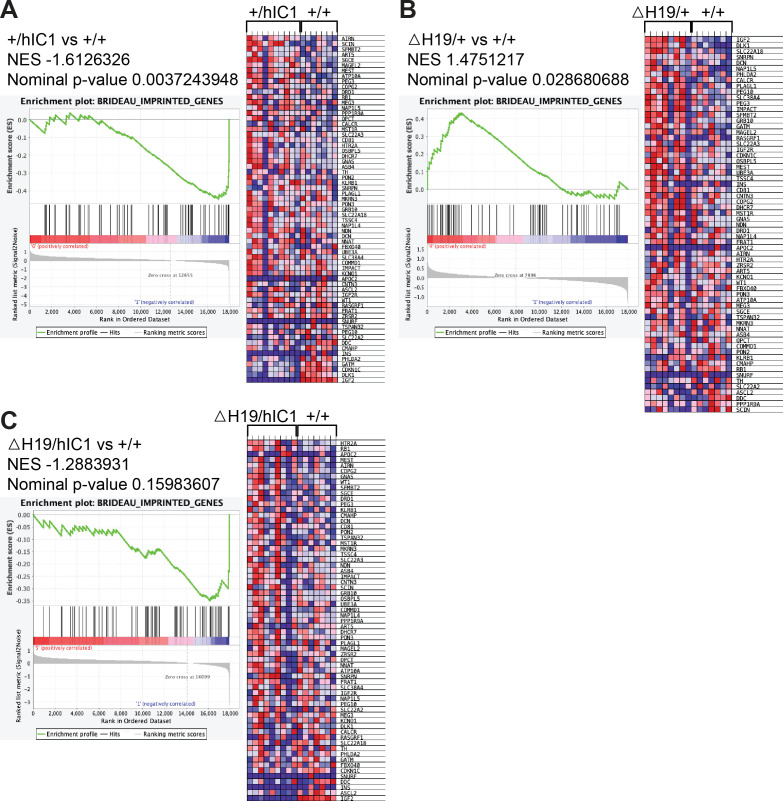
Figure 6. Transcriptomic analysis of E12.5 cardiac endothelial cells from wild-type and mutant embryos.
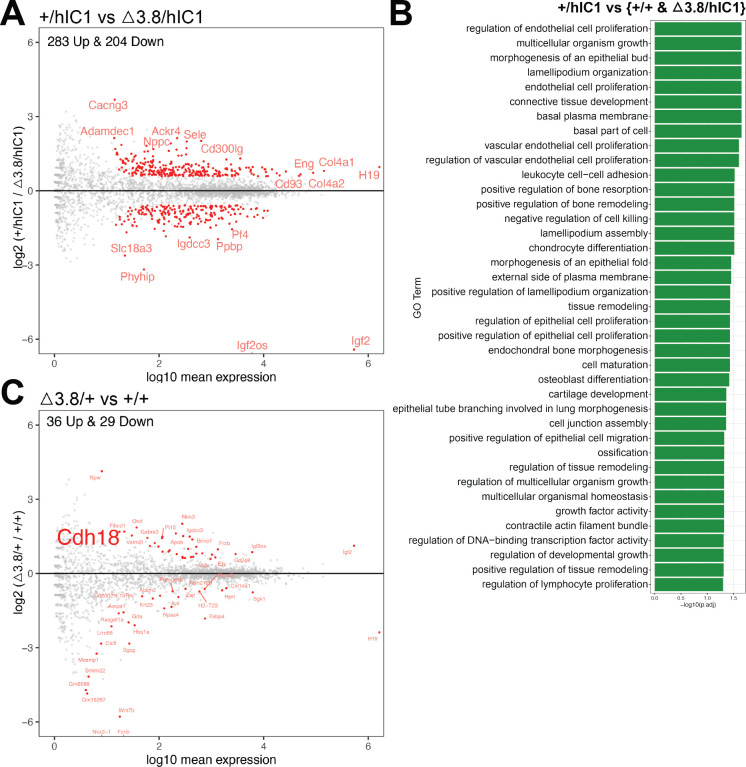
(A) A gradient H19 and Igf2 expression levels are depicted in E12.5 wild-type, +/hIC1, △H19/+, △H19/hIC1, △3.8/+, and △3.8/hIC1 cardiac endothelial cells. M: male and F: female. (B) Left: Comparison between +/hIC1 and the wild-type samples with a volcano plot shows 224 differentially expressed genes (DEGs) between +/hIC1 and the wild-type samples. Right: Gene ontology (GO) pathways that are enriched for the 224 DEGs. (C) Immunofluorescence staining for MF20 (red) and collagen (green) on E17.5 wild-type and +/hIC1 hearts. Images on the right are enlarged from the boxed area of images on the left. Scale bars = 100 µm. (D) Expression pattern of 25 genes that are differentially expressed in both male and female +/hIC1 samples is compared to wild-type. (E) A volcano plot represents two DEGs between △3.8/hIC1 and the wild-type samples. (F) Expression pattern of 116 genes that are commonly differentially expressed in +/hIC1 compared to the wild-type and △3.8/hIC1 samples.