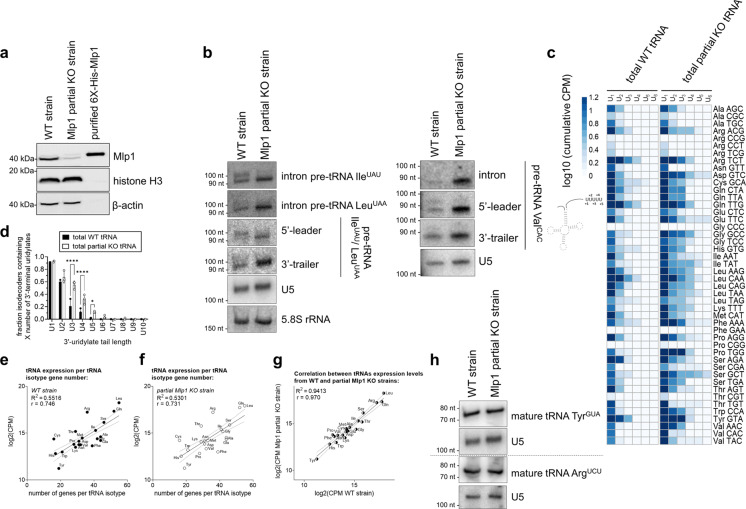
Fig. 5. Depletion of Mlp1 results in altered pre-tRNA processing in T. thermophila with normal levels of mature tRNAs.
a Western blot confirming decreased protein expression of Mlp1 in the partial Mlp1 knockout (KO) strain compared to the wild-type (WT) strain (n = 3 biologically independent samples). Loading control: histone H3 and β-actin. Purified 6X-His-Mlp1 was analyzed to control for antibody specificity. b Northern blot detecting tRNAs IleUAU, LeuUAA, and ValCAC pre-tRNA intermediates with an intron, 5’-leader, and 3’-trailer specific probe (n = 3 biologically independent samples). Pre-tRNA IleUAU and LeuUAA are both recognized by 3’-trailer and 5’-leader probe used. U5 snRNA and/or 5.8 S rRNA were used as loading controls. c Next-generation sequencing data shown as a heatmap of log10 transformed normalized cumulative pre-tRNA counts for each uridylate tail length (n = 3 biologically independent replicates). For clarity, the average is plotted and the longest uridylate tail shown is U6 (UUUUUU-3’OH). d Quantification of uridylate tail length from next-generation sequencing data (n = 3 biologically independent samples) in c. Error bars represent the mean +/− standard deviation. A two-way ANOVA with Bonferroni’s correction for multiple comparisons was performed to determine statistical significance: ****P < 0.0001, *P = 0.0491. For each tRNA isodecoder, the presence of a uridylate tail length (U1, U2, etc.) was scored as present if its abundance in CPM was greater than 0.3; log10(2). e, f Scatterplots of log2 transformed normalized tRNA counts (CPM) from next-generation sequencing data from WT strains (e) and partial Mlp1 KO strains (f) plotted against the number of genes per tRNA isotype encoded in the genome (see Supplementary Data 1). The correlation was calculated, and a positive correlation was observed for both WT and KO strains (r = 0.746 and r = 0.731, respectively) indicating that more tRNA gene copies result in higher tRNA expression. g Plotting of log2 transformed normalized WT and partial Mlp1 KO counts (CPM) gives a strong positive correlation (r = 0.970). h Northern blot analysis detecting mature tRNA TyrGUA and ArgUCU levels using a probe specific for the spliced mature tRNA (n = 3 biologically independent samples). U5 was used as a loading control on both membranes. Source data are provided as a Source Data file.