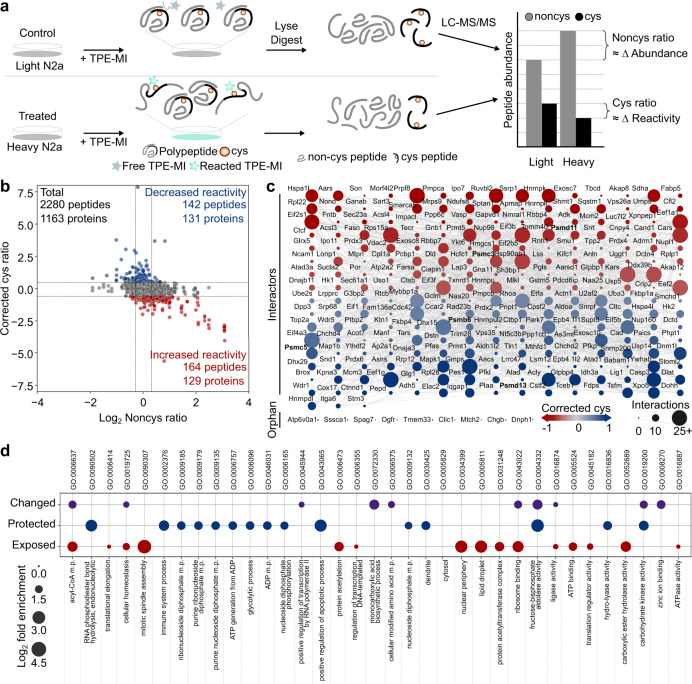
Fig. 2. Conformational changes due to proteasome inhibition reflect cellular responses to MG132.
a Method schematic for quantifying proteome conformation via proteomics. SILAC-labeled Neuro-2a cells were treated with pharmacological stimuli i.e., MG132 (heavy) or vehicle control DMSO (light), then labeled with TPE-MI and prepared for proteome analysis using LC-MS/MS. Peptide quantitation yielded the relative abundance of noncys-containing peptides (a measure of abundance) and cys-containing peptides (a measure of conformational status). b Representative scatterplot for processed noncys- and corrected cys-peptide abundance ratios in the presence of proteasome inhibitor MG132. Dots show per-peptide summary values across four biological replicates. Thresholds (red dotted lines) determined based on the control dataset are shown, outside which cysteine-containing peptides were considered more exposed (red) or more protected (blue). c Protein interaction network for changed peptides derived from b. Protein nodes are colored according to maximum corrected cys ratio and edges (lines) connect proteins with a known interaction (STRINGdb v 11.0, medium confidence score >0.4). Protein nodes are sized according to the number of interactions within the network. d Significantly enriched gene ontology terms (p < 0.05) for all proteins which changed reactivity (purple); or, more specifically, became protected (blue) or exposed (red) in b. Enrichment terms are filtered to minimize hierarchical redundancy (PantherGOSlim v 16.0). Source data are provided as a Source Data file.