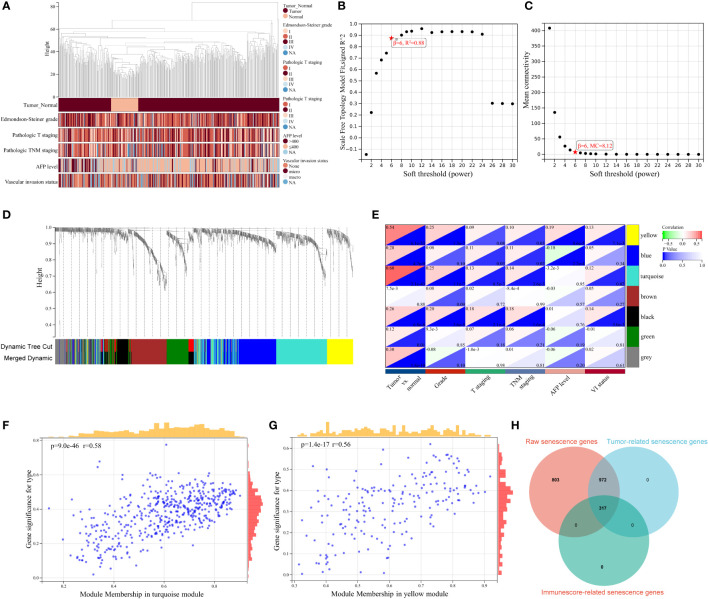
Figure 2.
WGCNA for identification of tumor-related senescence genes. (A) Clustering dendrogram of HCC samples and the clinical traits, covering sample type (tumor and normal), Edmondson–Steiner grade, pathologic T staging, pathologic TNM staging, alpha fetoprotein (AFP) level, and vascular invasion (VI) status. Soft threshold selection to determine the WGCNA module depending on scale independence (B) and mean connectivity (C). (D) Seven colored modules were determined and visualized with a dendrogram, based on a dissimilarity measure (1-TOM). (E) Heatmap for the correlation between gene modules and clinical characteristics. Scatter plots delineating the correlation between gene significance (GS) for tumor trait and module membership (MM) in the turquoise (F) and yellow module (G). (H) Venn diagram showing 317 overlapping tumorigenesis- and immune infiltration-associated genes. WGCNA, weighted gene co-expression network analysis; MC, mean connectivity; NA, not available.