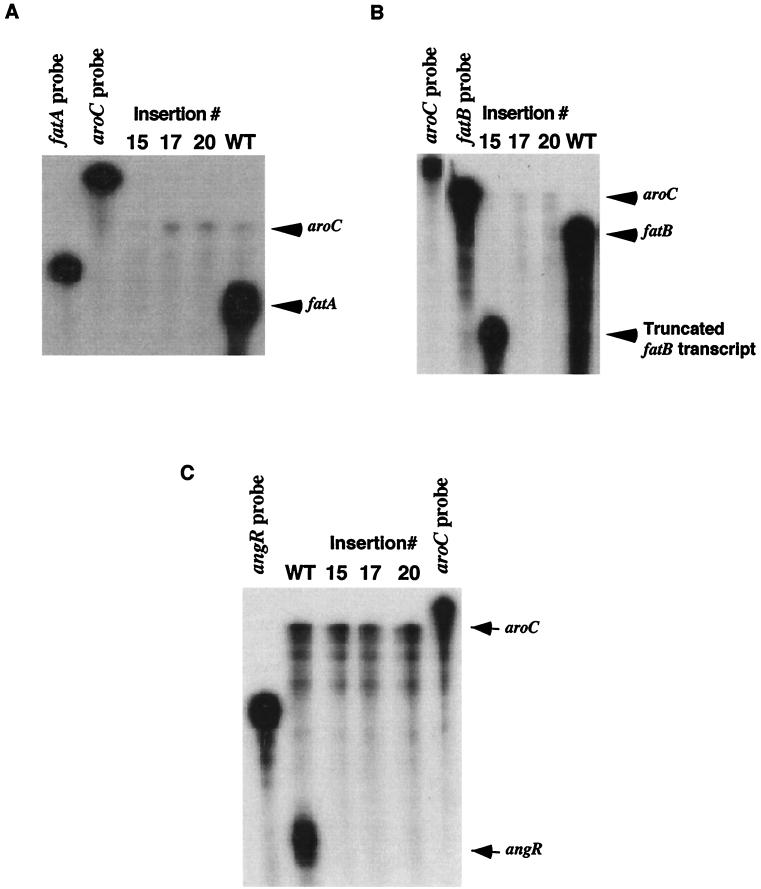
FIG. 2.
Effect of insertion mutations on transcription of the fatA, fatB, and angR genes as determined by RNase protection assays. Total RNA was harvested from V. anguillarum strains harboring various recombinant clones grown under iron-limiting conditions. Riboprobes were generated by using constructs pMET13.1, pJHC-LW260, and pQC3.5 (described in Table 1) and transcribed with T3 RNA polymerase, generating the fatA, fatB, and aroC probes, respectively. The aroC-specific riboprobe was included in the hybridization buffer as an internal control. For all three panels the specific transcripts were detected by RNase protection assays and are indicated by arrows, while the strains are indicated above each lane. Lanes marked aroC, fatB, fatA, or angR are the free riboprobes without RNase treatment. WT, TnangR4(pJHC-T2612); insertion 15, TnangR4(pJHC-T2612::TnfatB15); insertion 17, TnangR4(pJHC-T2612::TnfatC17); insertion 20, TnangR4(pJHC-T2612::TnfatD20). TnangR4 is V. anguillarum H775-3(pJHC-T2612::TnangR4, pJHC9-8). Tn is transposition sequence Tn3-HoHo1. Panels: A, detection of fatA-specific transcripts with the fatA probe; B, detection of fatB-specific transcripts with the fatB probe; C, detection of angR-specific transcripts with the angR probe.