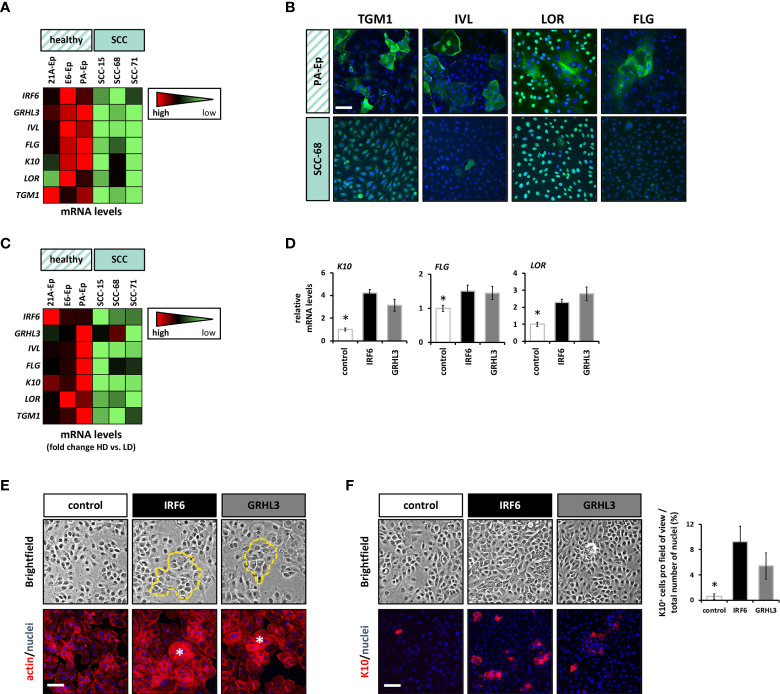
Figure 4.
Overexpression of IRF6 and GRHL3 induces differentiation. (A) The differentiation status of three normal cell strains compared to three SCC lines was assessed by qPCR for a panel of differentiation markers (IRF6, GRHL3, IVL, FLG, K10, LOR, and TGM1). The data are shown as a heatmap. Scale: light green – low gene expression; red – high gene expression. (B) IF staining for TGM1, IVL, LOR, and FLG in PA-Ep (non-cancerous) vs. SCC-68 indicates less differentiated cells in the SCC-68 cultures. Scale bar: 50 µm. Note that staining for LOR and TGM1 resulted in a nuclear background staining ( Supplementary Figure 5 ). (C) The same healthy cell strains and SCC cell lines were analyzed in low density (LD) and their corresponding high density (HD) cultures, and the same set of differentiation genes was analyzed by qPCR. The results are shown as heatmap of the fold inductions (HD vs. LD). Scale: light green – low gene induction; red – high gene induction. (D) The effect of forced expression of IRF6 (black) and GRHL3 (gray) compared to control SCC-68 (white) on differentiation markers (K10, FLG, and LOR) was determined by qPCR. * p < 0.05 control vs. IRF6 and GRHL3. (E) Brightfield pictures and corresponding pictures with actin staining indicate the presence of differentiating cell groups (dashed yellow line in the brightfield images and asterisks in the actin staining) in SCC68 cells transduced with IRF6 or GRHL3. Scale bar: 50 µm. (F) K10 staining in dense cultures of transduced SCC-68 cells (left) and its quantification (right). Note the presence of significantly more K10-positive cells in SCC-68/IRF6 and SCC-68/GRHL3 compared to SCC-68 control. Scale bar: 50 µm. * p < 0.05 control vs. IRF6 and GRHL3.