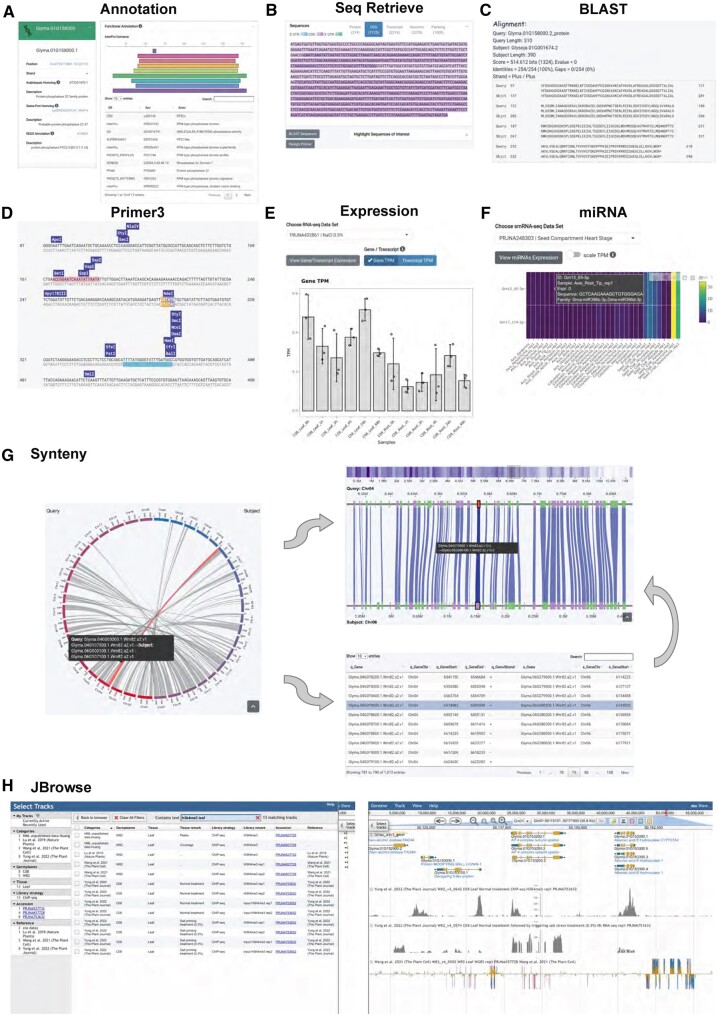
Figure 1.
Overview of the Wildsoydb DataHub web interface, using Glyma.01G158000 queried against the published soybean genome, Wm82 a4v1, as an example. A, Comprehensive annotation of the query gene, including the homolog in the Arabidopsis genome and Swiss-Prot, the annotation in the KEGG database (left), and functional annotations of the protein motif discovered by Interproscan (right). B, The DNA sequence of the query gene. C, Protein BLAST results. D, Primer3 result of the query gene. E, Expression levels of the query gene in a salt treatment RNA-seq dataset. F, Expressions of the miRNAs are predicted to target the query gene from a seed development dataset. G, Intra-genome synteny analysis of Wm82 a2v1. The macro-synteny blocks were illustrated with a circular layout (left), while the gene density and local micro-synteny regions were represented as a heatmap and in a parallel layout (right top). All the genes within the macro-synteny block were shown in a searchable table (right bottom). H, Jbrowse faceted track selector (left) and a demonstration view of the genomic regions around Glyma.01G153200 with an H3K4me3 ChIP-seq track, an RNA-seq track, as well as a BS-seq track on display (right; top to bottom).