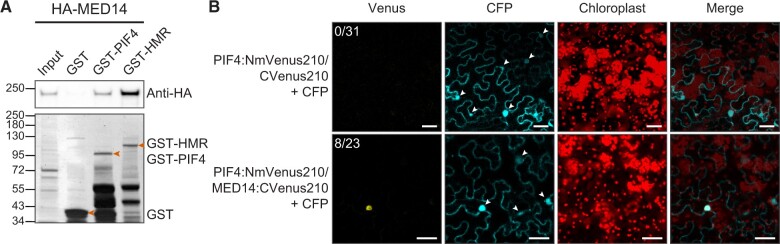
Figure 4.
MED14 physically interacts with PIF4 and HMR. (A) GST pull-down assays. GST-tagged full-length PIF4 and HMR were used as baits to pull down in vitro translated HA-tagged MED14. The upper panel is an immunoblot using anti-HA antibodies showing the bound and input fractions of HA-MED14; the lower part is a Coomassie Blue-stained SDS–PAGE gel showing immobilized GST and GST-tagged PIF4 and HMR. B, Bimolecular fluorescence complementation assays. PIF4 fused with NmVenus210 (PIF4:NmVenus210) and MED14 fused with cVenus210 (MED14:CVenus210) were co-expressed in N. benthamiana through agroinfiltration. PIF4:NmVenus210 and CVenus210 were co-expressed as a control. The CFP was expressed simultaneously through agroinfiltration to indicate the locations of nuclei. The optical density of each Agrobacterium strain was 0.1 and images were captured 40–70 h after infiltration. White arrowheads indicate nuclei identified in the CFP channel. The nuclear fluorescence signal in the Venus channel indicates the interactions between the two proteins tested in each agroinfiltration. The number of nuclei showing the Venus signal out of the number of nuclei with the CFP signal is shown in the upper left corner of each Venus channel. The chloroplast channel is to show that the Venus signal was not caused by the chlorophyll autofluorescence. Scale bars, 40 µm.