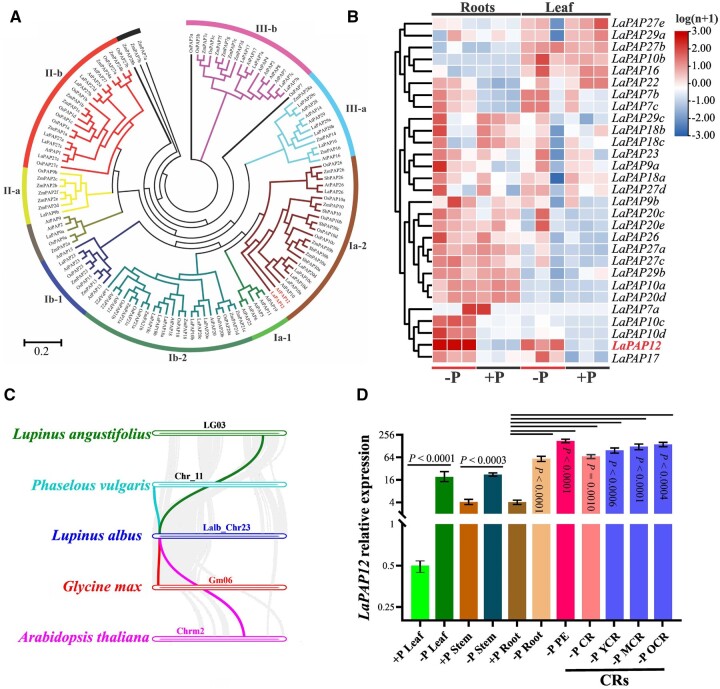
Figure 1.
Identification and characterization of white lupin PAPs. A, Phylogenetic analysis of PAPs in L. albus, A. thaliana, Z. mays, and O. sativa. The PAPs were aligned to generate an unrooted maximum-likelihood phylogenetic tree using IQ-TREE software, with 1,000 bootstrap replicates. B, Heatmap of the expression of white lupin PAPs in leaf and root grown under +P (soil with P fertilizer) and –P (soil without P fertilizer) conditions, based on the results of RT-qPCR. C, Results of a co-evolutionary analysis of PAP12 genes in L. albus, L. angustifolius, P. vulgaris, G. max, and A. thaliana, shown by the colored lines. D, Tissue/organ-specific expression of LaPAP12 in the leaf, stem, root, CR pre-emergence zone (PE), CRs, young CR (YCR), mature CR (MCR), and OCR of lupin grown under +P and–P hydroponic conditions. For two soil-P treatments: +P is 142.6 ppm (100 ppm P from P fertilizer + 42.6 ppm soil Olsen-P) and −P is 42.6 ppm (soil Olsen-P).