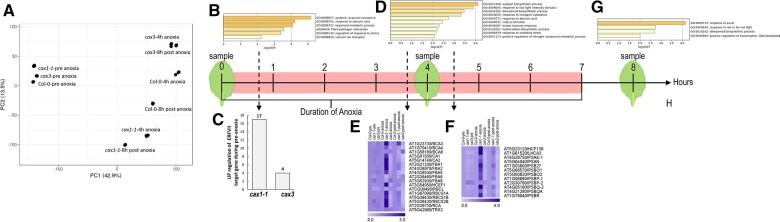
Figure 5.
CAX mutants transcriptome-wide responses to anoxia. A, PCA representation of Col-0, cax1, and cax3 transcriptomes before, during, and after anoxic treatment. B, GO groups identified in cax1 pre-anoxia that overlap with genes differentially expressed in col-0 and cax3 during anoxia (4-h) and post anoxia (1 h post 7-h anoxia). C, Bar graph denotes the upregulated HRGs downstream to ERFVII in pre-anoxic conditions. D, Substantially differential expression GO groups specifically found in cax1 after 4 h of anoxia. E, Preferential upregulation of genes involved in photosystem maintenance and cax1 following anoxia. F, Preferential upregulation of genes involved in carbohydrate metabolism in cax1 following anoxia. Fold change expression of the genes were used for constructing heat map using MeV_4_9_0 in both (c) and (F). G, Substantially differential expression GO groups specifically found in cax1 during post-anoxic conditions. H, Scheme and timing of leaf tissue harvesting for RNA-seq analysis. Plants belonging to nine rosette leaf stage were taken for RNA-seq analysis before anoxia (pre-conditions), after 4 h of anoxia (anoxia), and 1 h after a 7-h anoxia treatment (post anoxia).