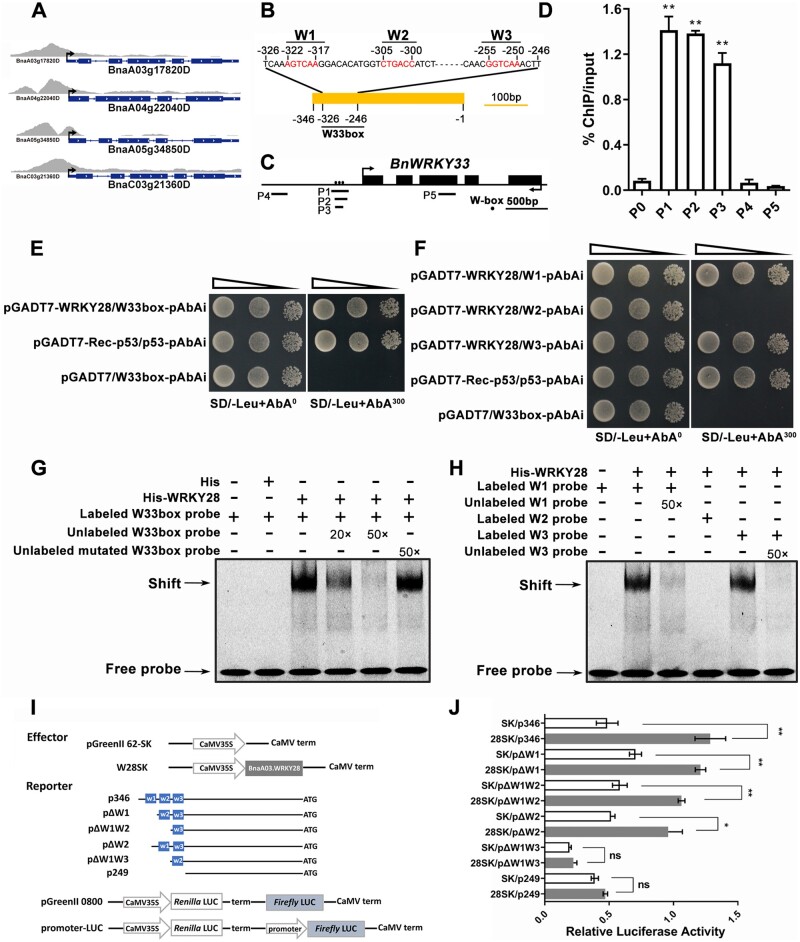
Figure 2.
BnaA03.WRKY28 binds to the BnWRKY33 promoter in vivo and in vitro. A, ChIP-seq showing specifically enrichment in the promoter regions of four copies of BnWRKY33. B, Characterization of the BnWRKY33 promoter sequence and the positions of W-boxes. The core sequence of the W-box is marked. C, A diagram of the BnWRKY33 locus showing the positions of amplicons (P1–P5) used for ChIP-qPCR. W-box elements are marked with circles. Amplicons: P1 (–370 to –166 bp), P2 (–330 to –181 bp), P3 (–328 to –233 bp), P4 (–1085 to –894 bp), and P5 (located in gene body). D, ChIP-qPCR to verify the binding of BnaA03.WRKY28 to the W33box region of BnWRKY33. The input and precipitated DNA were analyzed by qPCR primers. BnACTIN2 (P0), the promoter region distant from the start codon (P4), and the exon region of BnWRKY33 (P5) were used as negative controls. % input method was employed to determine the enrichment of BnaA03.WRKY28 at designated loci. Asterisks indicate significant differences compared with BnACTIN2 (t test; **P < 0.01). E and F, Y1H assays to explore the binding capacity of BnaA03.WRKY28 to the BnWRKY33 promoter. DNA fragments (W33box, –326 to –246 bp; W1/W2/W3, −326 to −313 bp, −309 to −296 bp, and −259 to −246 bp for three times tandem duplication, respectively) from the BnWRKY33 promoter were cloned into the pAbAi vector, and the CDS of BnaA03.WRKY28 was cloned into the pGADT7 vector (pGADT7-WRKY28). SD/-Leu, selective dropout medium without leucine. AbA300, medium containing 300 ng/mL AbA; AbA0, medium without AbA. p53 was used as a positive control. Oblique triangles above the images indicate a 10°, 10−1, and 10−2 yeast-cell concentration gradient. G and H, EMSA to test the interaction between BnaA03.WRKY28 and the BnWRKY33 promoter in vitro. Probes labeled with Cy5 were used. The mutated probe replaced the core W-box sequence (W1, W2, and W3) with GAGAGA. I, Schematic of effector and reporter constructs using the pGreenII vector set. The CDSs of the TFs were introduced into the pGreenII 62-SK (SK) vector as effectors. Truncated promoter regions of BnWRKY33 were inserted in front of the Firefly LUC as reporters. Renilla LUC was used as an internal control. J, Dual-LUC assays were conducted to detect relative LUC activity (the ratio of Firefly to Renilla LUC activity) when co-transformed with effector and reporter. Co-transformed empty pGreenII 62-SK plasmids and reporter constructs were used as negative controls. Data are shown as means ± sd (n = 3). Asterisks indicate significant differences compared with the negative controls (t test; ns, P > 0.05, *P < 0.05, **P < 0.01).