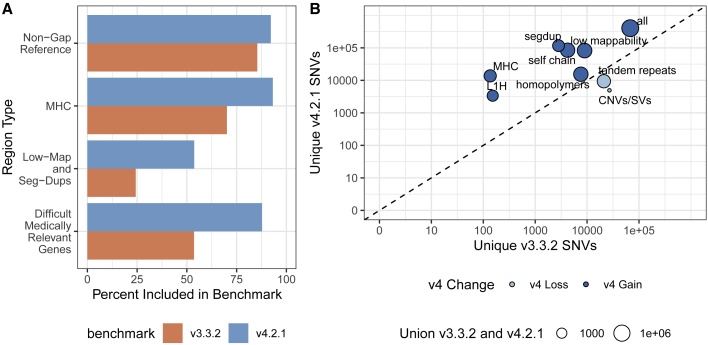
Figure 1.
The new benchmark set includes more of the reference genome and more variants
(A) Percentage of the genomic region that is included by HG002 v.3.3.2 and v.4.2.1 of all non-gap, autosomal GRCh38 bases; the MHC; low-mappability regions and segmental duplications; and 159 difficult-to-map, medically relevant genes described previously.
(B) The number of unique SNVs by genomic context. Circle size indicates the total number of SNVs in the union of v.3.3.2 and v.4.2.1. Circles above the diagonal indicate a net gain of SNVs in the newer benchmark, and circles below the diagonal indicate a net loss of SNVs in the newer benchmark.