Extended Data Fig. 3. Genetic abrogation of LSD1 impaired proliferation and survival of endocrine sensitive and resistant cells, and the LSD1 interactome.
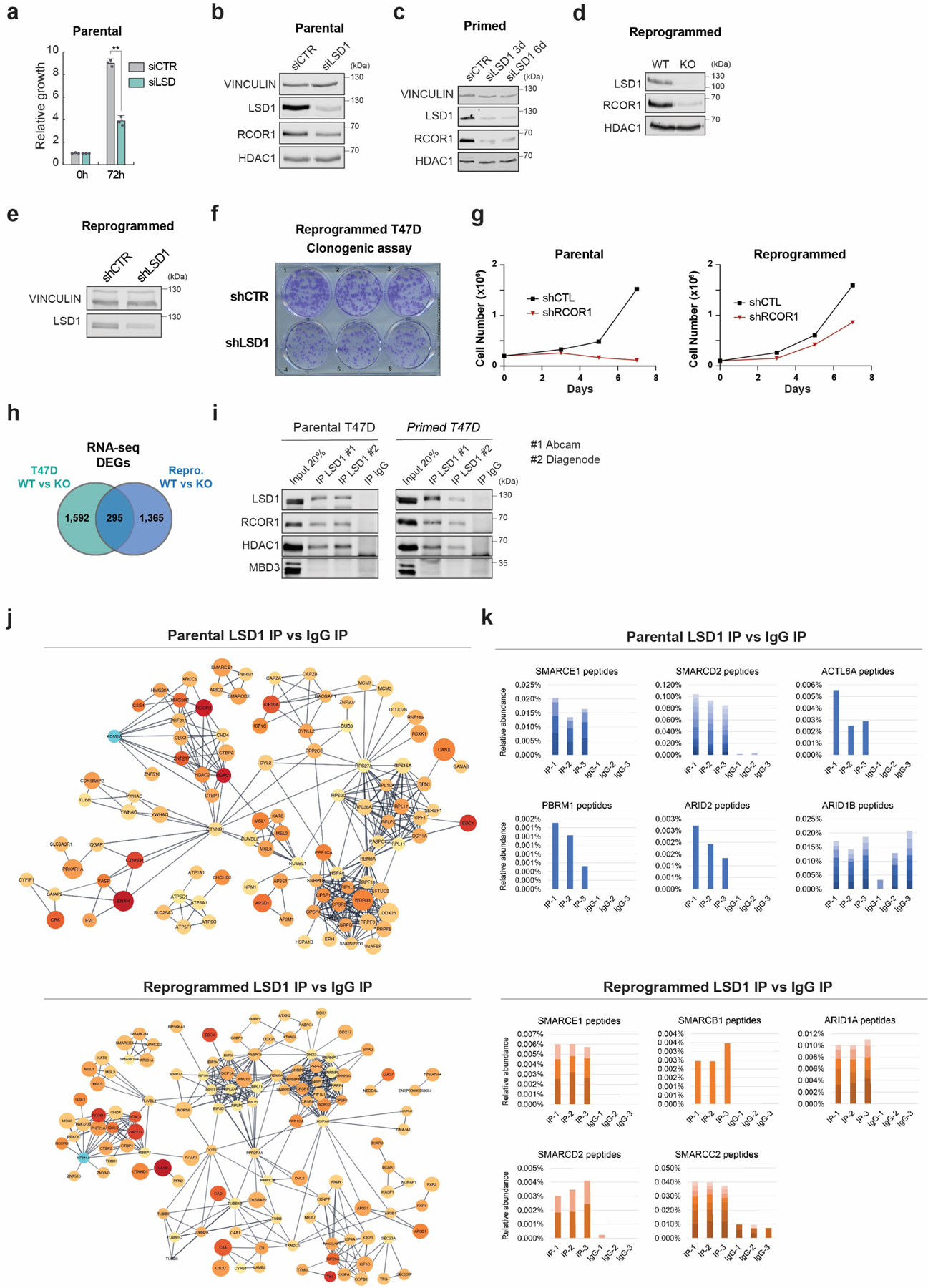
a, Proliferation of siLSD1 parental T47D 72h after siRNA transfection, n = 3 biological independent replicates. Data are presented as mean values + SEM. p-value < 0.005 (one-way ANOVA). b-e, WB of proteins indicated, with VINCULIN as the loading control, in siCTR and siLSD1 T47D 6 days after transfection (b), primed siLSD1 T47D 3- and 6-days post siRNA transfection (c) and in WT and KO reprogrammed T47D (d), and reprogrammed shCTR and shLSD1 T47D (e). f, Clonogenic assay of reprogrammed shLSD1 T47D, n = 3 biological independent replicates. g, Proliferation of shCTR and shRCOR1 parental and reprogrammed T47D for 7 days, n = 2 biological independent replicates. h, DEG overlap between parental and reprogrammed LSD1 KO T47D. i, Endogenous LSD1 immunoprecipitation (IP) with CoREST subunits in whole cell lysates using two antibodies in parental or primed T47D. IgG and MBD3 were used as negative controls. j, Interaction network of LSD1 interactome in parental and reprogrammed T47D. k, Relative peptide abundance of selected SWI/SNF subunits identified by LC-MS/MS in parental and reprogrammed T47D. IP = LSD1 IP, IgG = IgG IP. n = 3 biological independent replicates. Number of peptides are represented as shades of blue and orange. Uncropped images are available as source data.
