Fig. 5: The CoREST-cJUN axis in the reprogrammed cells.
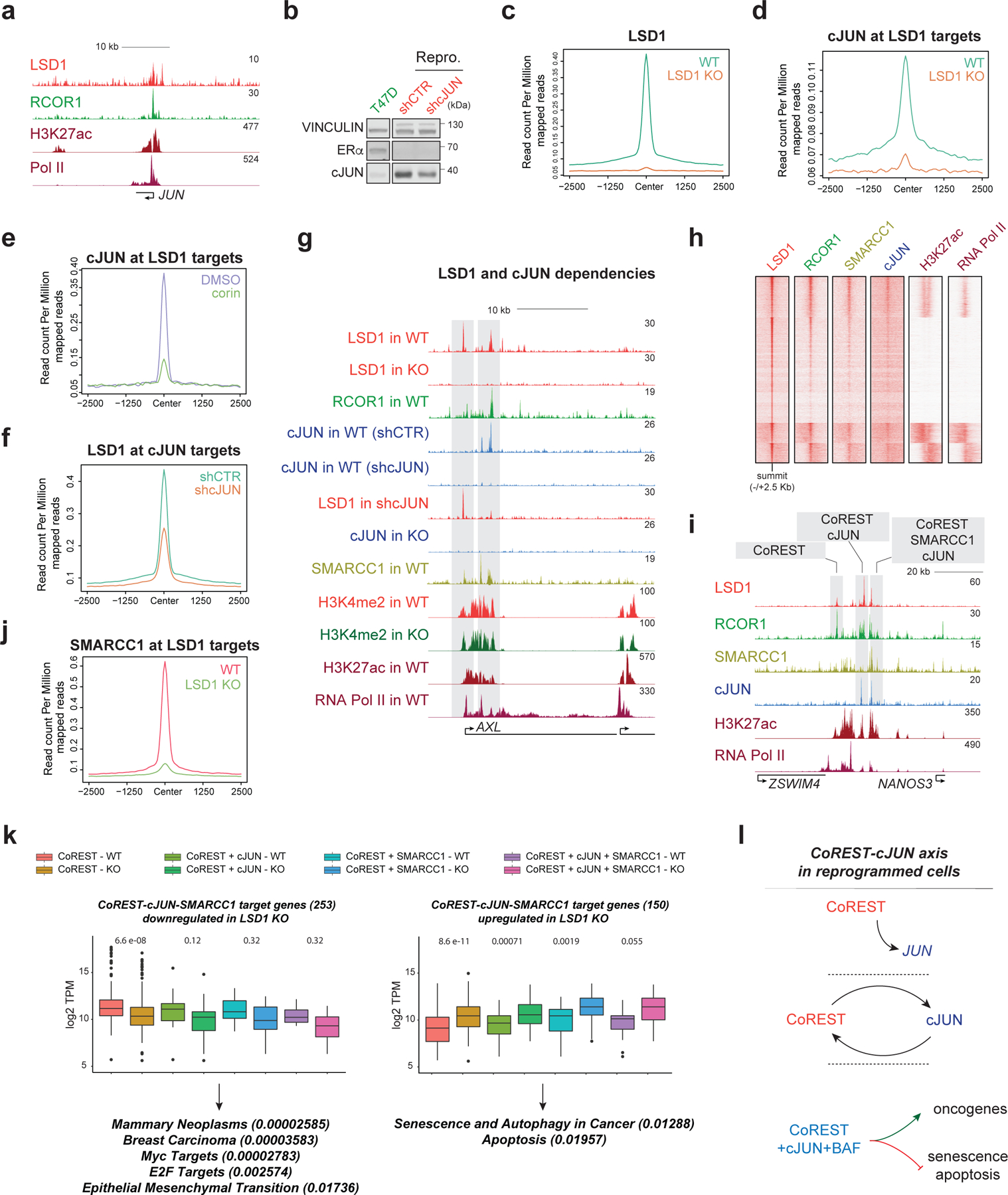
a, LSD1, RCOR1, H3K27ac, and RNA-Pol II ChIP-seq signal at the JUN promoter in reprogrammed cells. b, cJUN WB in parental and reprogrammed shCTR and shcJUN cells. c, LSD1 ChIP-seq signal in reprogrammed WT and LSD1 KO cells. d, cJUN ChIP-seq signal in reprogrammed WT and LSD1 KO cells. e, cJUN ChIP-seq signal at LSD1 targets in reprogrammed cells treated for 72h with 500nM corin. f, LSD1 ChIP-seq signal at cJUN targets in shCTR and shcJUN reprogrammed cells. g, ChIP-seq signal of factors indicated in WT and LSD1 KO reprogrammed cells and in shCTR and shcJUN cells at the AXL gene. h, ChIP-seq heatmap of factors indicated (23,979 peaks total) in reprogrammed cells. i, Co-occupancy profiles at the NANOS3 enhancer in reprogrammed cells. j, SMARCC1 ChIP-seq signal in reprogrammed WT and LSD1 KO cells. k, Downregulated (left) or upregulated (right) expression of CoREST target genes in reprogrammed WT and LSD1 KO cells. n=2 biologically independent samples. Downregulated genes were involved in EMT and more aggressive phenotypes while upregulated genes were associated with cell death, The box plots span from the 25th to 75th percentiles, the center line shows the median and whiskers show maximum and minimum values, p-values indicated in parentheses (Mann-Whitney two-sided test). l, Schematic of the CoREST-cJUN axis in reprogrammed cells. CoREST binds to the JUN promoter (top), CoREST-cJUN are recruited to chromatin in a co-dependent manner (middle), and CoREST-cJUN cooperate to activate genes (bottom). Uncropped images for b are available as source data.
