Figure 1.
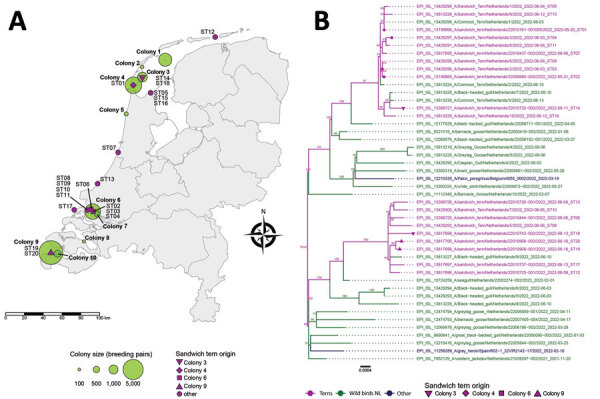
Location of Sandwich terns affected by locally acquired highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b viruses and phylogeny of viral segments, the Netherlands. A) Location and size (number of breeding pairs) of the Sandwich tern breeding colonies and the origin (finding location) of the Sandwich terns from which virus sequences ST01–ST20 shown in the phylogenetic tree in panel B were obtained. B) Maximum-likelihood tree (1,000 bootstraps) of the concatenated viral segments showing the H5N1 viruses detected in Sandwich terns together with viruses from other wild birds. Bootstrap values >50 are indicated at the branches. Identification numbers and symbols of the Sandwich terns correspond to those in the map, and the date that the bird was found dead is indicated. The GISAID sequences used in the phylogenetic analysis are listed in Appendix 1 Table 2. ST, Sandwich tern.
