Figure 1.
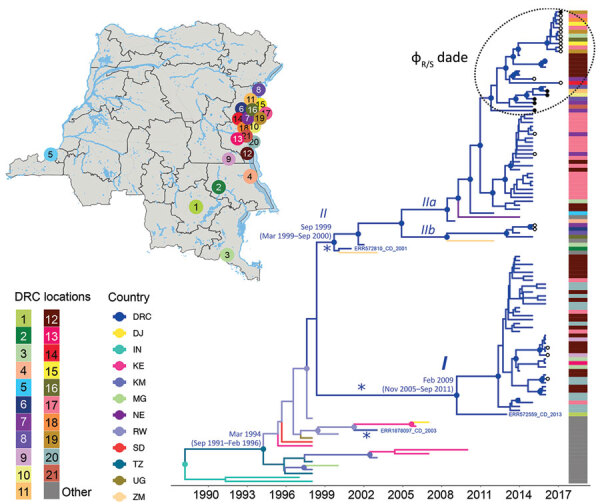
Spatiotemporal evolution and dissemination of Vibrio cholerae epidemic in the Democratic Republic of the Congo, 2015–2017. Sampling locations of V. cholerae strains sequenced in this study are indicated on the map. Each sampling location is coded by color and number, defined in the key; location colors are indicated for each tip in the maximum clade credibility tree as a heatmap (exact locations in Appendix Table 2). The tree was inferred from full genome V. cholerae isolates from DRC, neighboring countries, and Asia. Branches are scaled in time and colored by country of origin. Circles in internal nodes indicate posterior probability support >0.9, and the colors indicate ancestral countries inferred by Bayesian phylogeographic reconstruction. Circles at tips indicates the strains collected and sequenced in this study, with black circles designating phage-resistant strains. Notations I, II, IIa, and IIb indicate well-supported lineages and sublineages circulating in DRC during outbreaks. Asterisks (*) indicate potential spillover events within the Great Lakes region originated from neighboring countries. The tree with full tip labels is provided in Appendix Figure 2.
