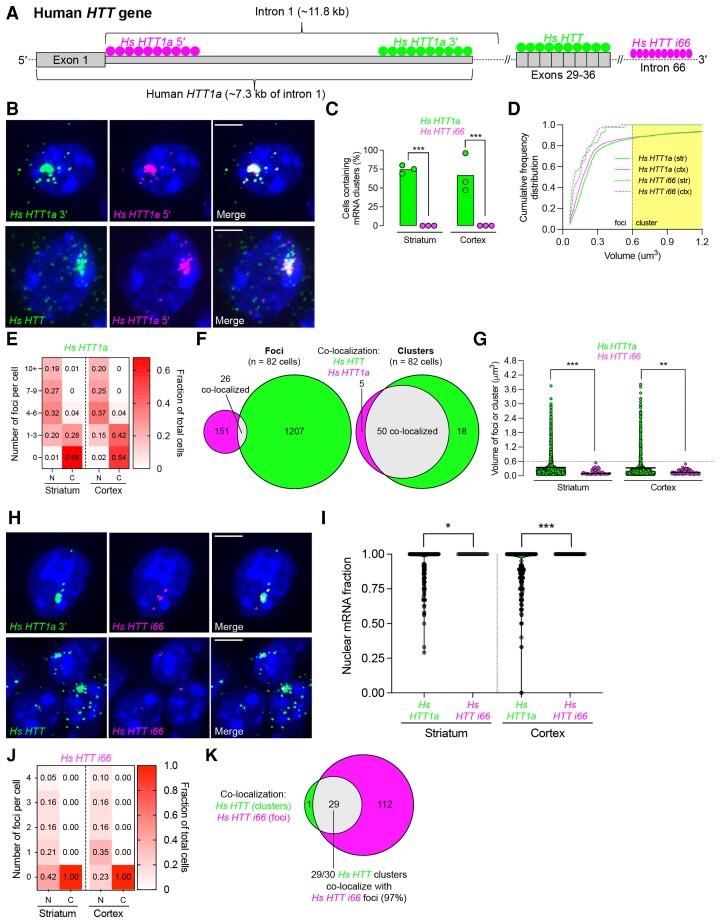
Figure 4.
Hs HTT1a, the aberrantly spliced exon 1-intron 1 fragment, is present in the cytoplasm and forms clusters that co-localize with Hs HTT clusters. (A) Gene schematic showing Hs HTT1a and Hs HTT i66. Filled circles indicate regions where FISH probes were designed. (B) Confocal microscope images of YAC128 mouse striatum (3 months old) detected by FISH. DAPI. Scale bar, 5 µm. (C) Percentage of cells containing Hs HTT1a or Hs HTT i66 mRNA clusters in YAC128 mouse striatum and cortex (n = ∼300 cells per brain region pooled from three mice, each point represents a mouse, one-way ANOVA with Tukey’s multiple comparisons test [F(3,8) = 29.20)]. (D) Cumulative frequency distribution plot of RNA foci volume. The yellow shaded area represents the cut-off for a cluster, which is defined to be at least 0.6 µm3. (E) Heatmap of the number of nuclear and cytoplasmic Hs HTT1a mRNA foci detected per individual cell by FISH. Each column adds up to 1. (F) Venn diagram depicting the co-localization of Hs HTT and Hs HTT1a mRNA analysed separately as foci versus clusters. (G) Scatter plot showing the volume of individual mRNA foci or clusters (see Methods for how volume was calculated). Each point represents the volume of individual mRNA foci and thick line represents the mean (Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test). (H) Same as (B) using different RNAscope probes. (I) Nuclear fraction of Hs HTT1a and Hs HTT i66 mRNA is in the striatum and cortex. Each point represents a cell (n = ∼300 cells pooled from three mice per brain region, Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test). (J) Same as (E), but looking at Hs HTT i66. (K) Same as (F), but looking at the co-localization of Hs HTT clusters and Hs HTT i66 foci. For all panels, ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.