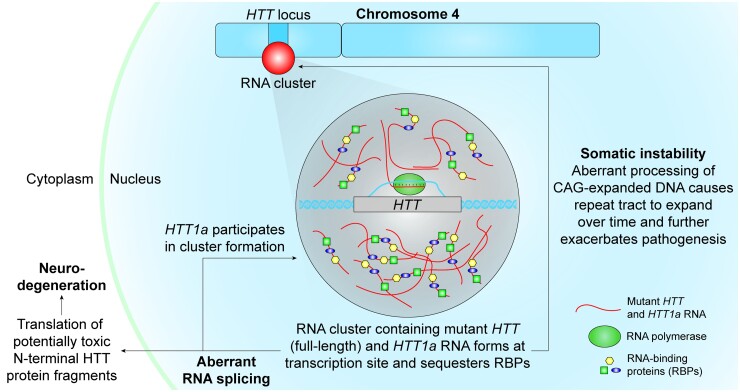
Figure 7.
Proposed model of mutant HTT RNA cluster formation, pathology, and pathogenesis. RNA clusters containing mutant HTT RNA nucleate at active transcription sites. These repeat-expanded RNAs sequester RBPs, removing them from the available cellular pool, and thus, disrupting downstream RNA processing such as splicing. HTT itself is aberrantly spliced to produce HTT1a, which also participates in cluster formation. Globally disrupted splicing can result in the translation of altered protein isoforms and lead to neurotoxicity. This entire process is exacerbated by somatic instability, which acts as a positive feedback loop and expands the CAG repeat tract in the HTT gene over time and further increases the rate of HTT1a production.